| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
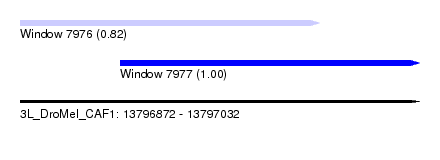
| Location | 13,796,872 – 13,797,032 |
| Length | 160 |
| Max. P | 0.999203 |

| Location | 13,796,872 – 13,796,992 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13796872 120 + 23771897 UAGUUUCACUAAGCACAACUAUUACAGAGUUUUCAAAUUGUAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAA ..((((.(((...(((.......(((..((((....(((((....)))))...))))...)))......((((((((((.....)))))).))))......)))...)))...))))... ( -19.30) >DroSec_CAF1 33769 120 + 1 UAGUUUCACUAAGCACAACUAUUACACAGUUUUCAAAUUGUAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAA ..((((.(((...(((............((((....(((((....)))))...))))...((((.....((((((((((.....)))))).))))))))..)))...)))...))))... ( -18.50) >DroSim_CAF1 29877 120 + 1 UAGUUUCACUAAGCACAACUAUUACACAGUUUUCAAAUUGUAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAA ..((((.(((...(((............((((....(((((....)))))...))))...((((.....((((((((((.....)))))).))))))))..)))...)))...))))... ( -18.50) >DroEre_CAF1 34872 119 + 1 UAGUUUCACUAAGCACAACUAUUACACAGUUUUCAAAUUGUAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUG-CACAACCAACGUGAGGAGUUAAAAACAAA ..((((.(((...(((............((((....(((((....)))))...))))............((((((((((.....))))))-))))......)))...)))...))))... ( -22.10) >DroYak_CAF1 32465 120 + 1 UAGUUUCACUAAGCACAACUAUUACACAGUUUUCAAAUUGUAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGCUAAAAACAAA (((((((..................((((((....)))))).(((....(((((......)))))....((((((((((.....)))))).)))).....)))..)))))))........ ( -19.90) >consensus UAGUUUCACUAAGCACAACUAUUACACAGUUUUCAAAUUGUAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAA ..((((.(((...(((.......(((..((((....(((((....)))))...))))...)))......((((((((((.....)))))).))))......)))...)))...))))... (-17.74 = -17.94 + 0.20)



| Location | 13,796,912 – 13,797,032 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -20.94 |
| Consensus MFE | -20.04 |
| Energy contribution | -19.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13796912 120 + 23771897 UAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAAGACAGCACAUUUACGACCACAAUAACAAUUGCAUUAUCAC .........((((((.(((((((......((((((((((.....)))))).))))(((..(....)..)))...............)))))))......((((....))))..)))))). ( -19.90) >DroSec_CAF1 33809 120 + 1 UAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAAGACAGCACAUUUACGACCACAAUAACAAUUGCAUUAUCAC .........((((((.(((((((......((((((((((.....)))))).))))(((..(....)..)))...............)))))))......((((....))))..)))))). ( -19.90) >DroSim_CAF1 29917 120 + 1 UAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAAGACAGCACAUUUACGACCACAAUAACAAUUGCAUUAUCAC .........((((((.(((((((......((((((((((.....)))))).))))(((..(....)..)))...............)))))))......((((....))))..)))))). ( -19.90) >DroEre_CAF1 34912 119 + 1 UAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUG-CACAACCAACGUGAGGAGUUAAAAACAAAGGCAGCACAUUUACGACCACAAUAACAAUUGCAUUAUCAC .........((((((.(((.(((((....((((((((((.....))))))-)))).....((((((..(((............)))...)))))).......))))).)))..)))))). ( -23.20) >DroYak_CAF1 32505 120 + 1 UAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGCUAAAAACAAAGGCAGCACAUUUACGACCACAAUAACAAUUGCAUUAUCAC .........((((((.(((.(((((....((((((((((.....)))))).)))).....((((((..(((............)))...)))))).......))))).)))..)))))). ( -21.80) >consensus UAACGACAAUGAUAAAUAAAUGUUAACAAUGUGCAAAUUGUUUAAAUUUGCCACAAACAACGUGAGGAGUUAAAAACAAAGACAGCACAUUUACGACCACAAUAACAAUUGCAUUAUCAC .........((((((.(((.(((((....((((((((((.....)))))).)))).....((((((..(((............)))...)))))).......))))).)))..)))))). (-20.04 = -19.88 + -0.16)

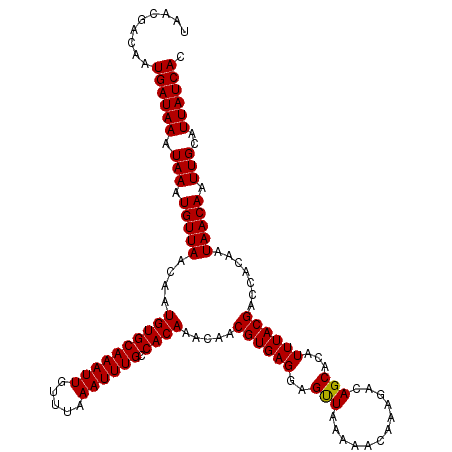

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:19 2006