
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,795,461 – 13,795,674 |
| Length | 213 |
| Max. P | 0.967827 |

| Location | 13,795,461 – 13,795,581 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13795461 120 - 23771897 ACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGGCGACGAAGACUUAAAACGUCGUUCGACAUAAAUCCCACCC .((((....)))).((((((............))))))....(((.((((....)))))))((((..((((....))))((((((...........))))))............)))).. ( -36.20) >DroSec_CAF1 32516 120 - 1 ACUGCAUAAGCAGAAGUGCCCCUAUCUAAAUAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGGCGACGAAGACUUAAAACGUCGUUCGACAUAAAUCCCACCC .((((....)))).((((((............))))))....(((.((((....)))))))((((..((((....))))((((((...........))))))............)))).. ( -36.20) >DroSim_CAF1 28645 120 - 1 ACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGGCGACGAAGACUUAAAACGUCGUUCGACAUAAAUCCCACCC .((((....)))).((((((............))))))....(((.((((....)))))))((((..((((....))))((((((...........))))))............)))).. ( -36.20) >DroEre_CAF1 33564 120 - 1 ACUGCAUAAGCAGAAGUGCCCCUAUCUAAGCAGCCACUAUAUAGAAUUGGUAGCCGAGUCUGUGGAACAGCUGUUGCUGGCGACGAAGACUUAAAACGUCGUUCGACAUAAAUCCCACCC .((((....))))...((((........((((((.....((((((.((((...)))).)))))).....))))))...)))).(((((((.......))).))))............... ( -29.80) >consensus ACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGGCGACGAAGACUUAAAACGUCGUUCGACAUAAAUCCCACCC .((((....)))).((((((............)))))).........(((((.....(((.......((((....))))((((((...........))))))..))).....)))))... (-32.30 = -32.80 + 0.50)



| Location | 13,795,501 – 13,795,594 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 97.31 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13795501 93 + 23771897 CCAGCAACAGCUGUUCCACAGACUGGGCUCCCAAUUCUAUAUAGUGCCUGUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCG .......((((((((.((.(((.(((....)))..)))....(((((((.(......).))))))).((((....)))).)).)))))))).. ( -31.50) >DroSec_CAF1 32556 93 + 1 CCAGCAACAGCUGUUCCACAGACUGGGCUCCCAAUUCUAUAUAGUGCCUAUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCG ..((((.((((((.....))).))).(((((....((((.((((...)))).))))..))))).....))))...(((((((...))))))). ( -30.30) >DroSim_CAF1 28685 93 + 1 CCAGCAACAGCUGUUCCACAGACUGGGCUCCCAAUUCUAUAUAGUGCCUGUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCG .......((((((((.((.(((.(((....)))..)))....(((((((.(......).))))))).((((....)))).)).)))))))).. ( -31.50) >DroEre_CAF1 33604 93 + 1 CCAGCAACAGCUGUUCCACAGACUCGGCUACCAAUUCUAUAUAGUGGCUGCUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCG .......((((((((((.......(((((((............))))))).........))(((...((((....)))).))))))))))).. ( -27.89) >consensus CCAGCAACAGCUGUUCCACAGACUGGGCUCCCAAUUCUAUAUAGUGCCUGUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCG .......((((((((.((........(((((....((((.((((...)))).))))..)))))....((((....)))).)).)))))))).. (-27.29 = -27.60 + 0.31)



| Location | 13,795,501 – 13,795,594 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 97.31 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
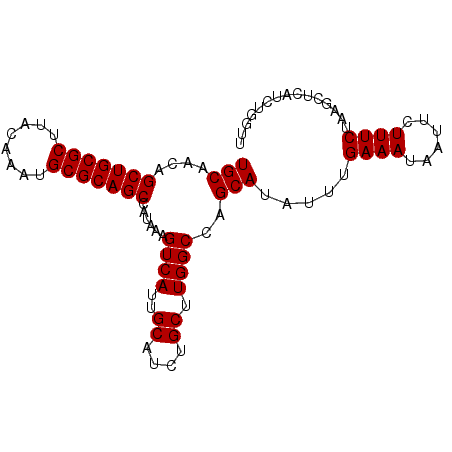
>3L_DroMel_CAF1 13795501 93 - 23771897 CGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGG .(((((((((.(..((((....)))).((((((............))))))..(((((.((((....)))))))))).)))))))))...... ( -36.20) >DroSec_CAF1 32556 93 - 1 CGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAAUAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGG .(((((((((.(..((((....)))).((((((............))))))..(((((.((((....)))))))))).)))))))))...... ( -36.20) >DroSim_CAF1 28685 93 - 1 CGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGG .(((((((((.(..((((....)))).((((((............))))))..(((((.((((....)))))))))).)))))))))...... ( -36.20) >DroEre_CAF1 33604 93 - 1 CGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAGCAGCCACUAUAUAGAAUUGGUAGCCGAGUCUGUGGAACAGCUGUUGCUGG .((((((((((((.((((....))))...)))(((...((..((.((((...........)))).))..))...).)))))))))))...... ( -29.40) >consensus CGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAUAUAGAAUUGGGAGCCCAGUCUGUGGAACAGCUGUUGCUGG .(((((((((.(..((((....)))).((((((............))))))..(((((.((((....)))))))))).)))))))))...... (-32.20 = -32.70 + 0.50)


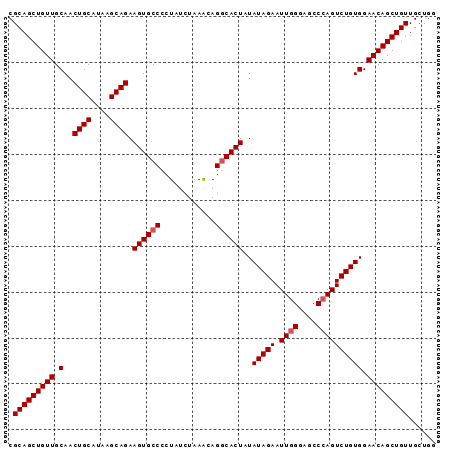
| Location | 13,795,541 – 13,795,634 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 97.85 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -32.05 |
| Energy contribution | -32.55 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13795541 93 + 23771897 AUAGUGCCUGUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGG ..(((((((.(......).)))))))...((..(((((((.((....(((((((........)))))))......)).)))))))..)).... ( -35.70) >DroSec_CAF1 32596 93 + 1 AUAGUGCCUAUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGG ..(((((((..........)))))))...((..(((((((.((....(((((((........)))))))......)).)))))))..)).... ( -34.00) >DroSim_CAF1 28725 93 + 1 AUAGUGCCUGUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGG ..(((((((.(......).)))))))...((..(((((((.((....(((((((........)))))))......)).)))))))..)).... ( -35.70) >DroEre_CAF1 33644 93 + 1 AUAGUGGCUGCUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCCGCUUGG ..(((((.(((...(((.(..(((...((((....)))).)))..).(((((((........)))))))......)))...))).)))))... ( -36.40) >consensus AUAGUGCCUGUUUAGAUAGGGGCACUUCUGCUUAUGCAGUUGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGG ..(((((((.(......).)))))))...((..(((((((.((....(((((((........)))))))......)).)))))))..)).... (-32.05 = -32.55 + 0.50)

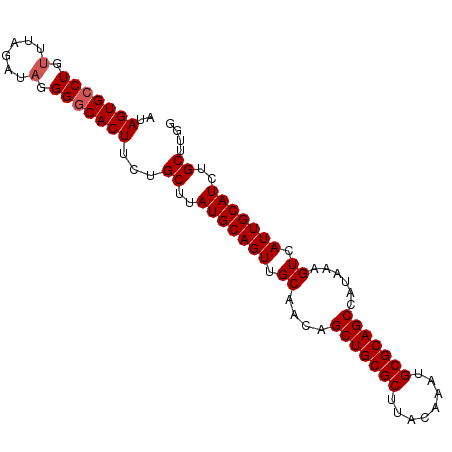
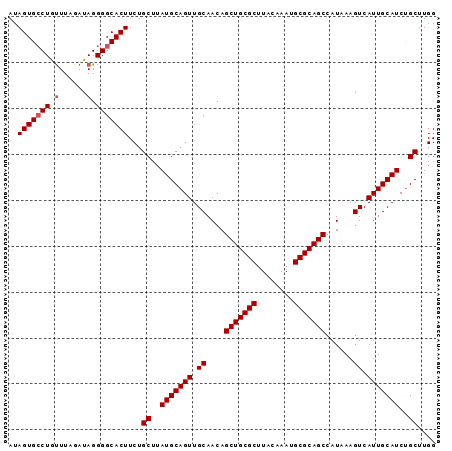
| Location | 13,795,541 – 13,795,634 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 97.85 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -33.39 |
| Energy contribution | -33.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13795541 93 - 23771897 CCAAGCAGAUGCAAUGACUUUAUGGCUGCGCAUUUGUAAGCGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAU ....((((.((((((........((((((((........)))))))))))))).)))).........((((((............)))))).. ( -35.30) >DroSec_CAF1 32596 93 - 1 CCAAGCAGAUGCAAUGACUUUAUGGCUGCGCAUUUGUAAGCGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAAUAGGCACUAU ....((((.((((((........((((((((........)))))))))))))).)))).........((((((............)))))).. ( -35.30) >DroSim_CAF1 28725 93 - 1 CCAAGCAGAUGCAAUGACUUUAUGGCUGCGCAUUUGUAAGCGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAU ....((((.((((((........((((((((........)))))))))))))).)))).........((((((............)))))).. ( -35.30) >DroEre_CAF1 33644 93 - 1 CCAAGCGGAUGCAAUGACUUUAUGGCUGCGCAUUUGUAAGCGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAGCAGCCACUAU ...((.((.(((.........((((((((((........)))))))))).(((.((((....))))...)))..........))).)).)).. ( -31.20) >consensus CCAAGCAGAUGCAAUGACUUUAUGGCUGCGCAUUUGUAAGCGCAGCUGUUGCAACUGCAUAAGCAGAAGUGCCCCUAUCUAAACAGGCACUAU ....((((.((((((........((((((((........)))))))))))))).)))).........((((((............)))))).. (-33.39 = -33.45 + 0.06)


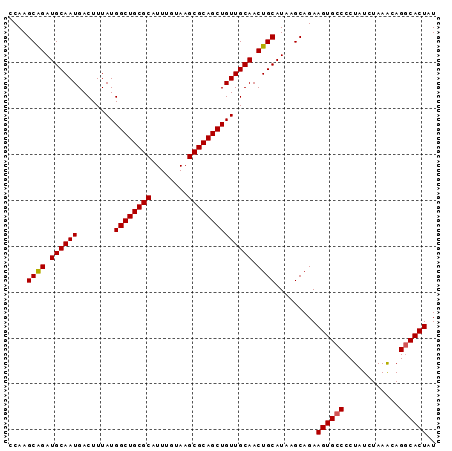
| Location | 13,795,581 – 13,795,674 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 99.46 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 1.00 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13795581 93 + 23771897 UGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGGCCAGCAUAUUUGAAAUAAUUCUUUCUAAGCUCAUCUGGUU (((....(((((((........)))))))......((((..((....)).))))..))).....((((......))))............... ( -22.10) >DroSec_CAF1 32636 93 + 1 UGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGGCCAGCAUAUUUGAAAUAAUUCUUUCUAAGCUCAUCUGGUU (((....(((((((........)))))))......((((..((....)).))))..))).....((((......))))............... ( -22.10) >DroSim_CAF1 28765 93 + 1 UGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGGCCAGCAUAUUUGAAAUAAUUCUUUCUAAGCUCAUCUGGUU (((....(((((((........)))))))......((((..((....)).))))..))).....((((......))))............... ( -22.10) >DroEre_CAF1 33684 93 + 1 UGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCCGCUUGGCCAGCAUAUUUGAAAUAAUUCUUUCUAAGCUCAUCUGGUU (((....(((((((........)))))))......((((..((....)).))))..))).....((((......))))............... ( -21.80) >consensus UGCAACAGCUGCGCUUACAAAUGCGCAGCCAUAAAGUCAUUGCAUCUGCUUGGCCAGCAUAUUUGAAAUAAUUCUUUCUAAGCUCAUCUGGUU (((....(((((((........)))))))......((((..((....)).))))..))).....((((......))))............... (-22.02 = -22.02 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:16 2006