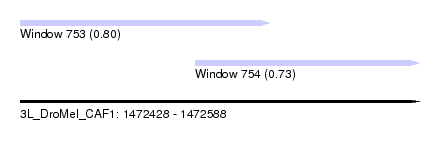
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,472,428 – 1,472,588 |
| Length | 160 |
| Max. P | 0.802706 |

| Location | 1,472,428 – 1,472,528 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.35 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -21.09 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
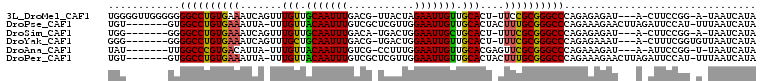
>3L_DroMel_CAF1 1472428 100 + 23771897 UGGGGUUGGGGGGGCCUGUGAAAUCAGUUUGUUGCAAUUUGACG-UUACUAGAAUUGUUGCACU-UUCCGCGGGCCCAGAGAGAU---A-CUUCCGG-A-UAAUCAUA ....(((.((.(((((((((.((..(((..((.(((((((....-......))))))).)))))-)).))))))))).(((....---.-))))).)-)-)....... ( -26.50) >DroPse_CAF1 159573 99 + 1 UGU-------GUGGCCUGUGAAAUUA-UUUGUUACAAUUUGUCGCUCGUUGGAAUUGUUGCACUACUUUGCGGGCCCAGAAAGAACUUAGAUUCCAU-UUUAAUCAUA (.(-------(.((((((..((....-..(((.(((((((..(....)...))))))).)))....))..)))))))).).........((((....-...))))... ( -21.60) >DroSim_CAF1 117510 93 + 1 UGG-------GGGGCCUGUGAAAUCAGUUUGUUGCAAUUUGACA-UGACUGGAAUUGCUGCACU-UUUCGCGGGCCCAGAGAGAU---A-CUUCCGG-A-UAAUCAUA ..(-------(((((((((((((..(((..((.(((((((.(..-....).))))))).)))))-)))))))))))).(((....---.-)))))..-.-........ ( -30.70) >DroYak_CAF1 125554 95 + 1 GGG-------GGGGCCUGUGAAAUCAGUUUGCUGCAAUUUGACG-UGACUGGAAUUGUUGCACU-UUUCGCGGGCCCAGAGAAAU---A-CUUUCGGUGUUAAUCAUA ...-------.((((((((((((..(((..((.(((((((.(..-....).))))))).)))))-)))))))))))).((..(((---(-(.....)))))..))... ( -34.60) >DroAna_CAF1 127678 93 + 1 UAU-------UUGGCCCGUGACAUUA-UUUGUUACAAUUUGUCG-CCUUUGGAAUUGUUGCACGAGUUCGCGGGCCCAGAAAGAU---A-AUUCCGG-U-UAAUCAUA ...-------..(((((((((.(((.-..(((.(((((((....-......))))))).)))..))))))))))))......(((---(-((....)-)-).)))... ( -24.70) >DroPer_CAF1 162193 99 + 1 UGU-------GUGGCCUGUGAAAUUA-UUUGUUACAAUUUGUCGCUCGUUGGAAUUGUUGCACUACUUUGCGGGCCCAGAAAGAACUUAGAUUCCAU-UUUAAUCAUA (.(-------(.((((((..((....-..(((.(((((((..(....)...))))))).)))....))..)))))))).).........((((....-...))))... ( -21.60) >consensus UGG_______GGGGCCUGUGAAAUCA_UUUGUUACAAUUUGACG_UCACUGGAAUUGUUGCACU_UUUCGCGGGCCCAGAAAGAU___A_AUUCCGG_U_UAAUCAUA ............((((((((((.......(((.(((((((...........))))))).)))....))))))))))................................ (-21.09 = -20.53 + -0.55)

| Location | 1,472,498 – 1,472,588 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.27 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -12.81 |
| Energy contribution | -12.92 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
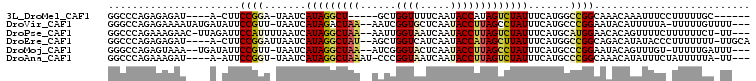
>3L_DroMel_CAF1 1472498 90 + 23771897 GGCCCAGAGAGAU----A-CUUCCGGA-UAAUCAUAGGCU-----GCUGGUUUUCAAUACCAUAGUCUAUUUCAUGGCCGGCAAACAAAUUUCCUUUUUGC------ ((((..(((((((----.-........-..))).((((((-----(.((((.......)))))))))))))))..)))).(((((...........)))))------ ( -21.30) >DroVir_CAF1 129462 100 + 1 GGGCCAGAGAAAAUAUGAUAUUCCGUU-UAAUCAUAGACUAA--AAUCGGUGCUCAAUACCUUAGCCUAUUUCAUGCCCGGAAUACAUUUUUA-UUUUUGUUUU--- ....(((((((((.(((.(((((((..-((...((((.((((--....((((.....)))))))).))))....))..)))))))))).))).-))))))....--- ( -18.80) >DroPse_CAF1 159637 100 + 1 GGCCCAGAAAGAAC-UUAGAUUCCAUUUUAAUCAUAGGCUAA--AAUUGGUAAUCAAUACCUUAGUCUAUUUCAUGCAUGGAACACAGUUUUCUUUUUUCU-UU--- .....(((((((((-(....((((((.......(((((((((--....((((.....))))))))))))).......))))))...)))..)))))))...-..--- ( -20.24) >DroEre_CAF1 124776 99 + 1 GGCCCAGAGAGAU----A-CUUCCGGAUUAAUCAUAGGCUAU--AGCUGGUCAUCAAUACCAUAGCUUAUUUCAUGGCCGGCAGACAUAUACCCUUUUUUU-UUGCA ((((..(((((((----.-...........))).(((((((.--...((((.......)))))))))))))))..)))).(((((...............)-)))). ( -19.96) >DroMoj_CAF1 107582 98 + 1 GGGCCAGAGUAAA--UGAUAUUCCGUU-UAAUCAUAGGCUAA--AUCGGGUACUCAAUACCUUAGCCUAUUUCAUGCCCGGAAUACAGUUUGU-UUUUUGAUUU--- ....(((((((((--(..(((((((..-((...((((((((.--...(((((.....)))))))))))))....))..)))))))..))))).-.)))))....--- ( -26.00) >DroAna_CAF1 127741 96 + 1 GGCCCAGAAAGAU----A-AUUCCGGU-UAAUCAUAGGCUAAAU-CCCGGUAAUCAAUACCUUAGUCUAUUUCAUGCCCGGCAAACAUAUUUCUAUUUUUA-UU--- .....(((((...----.-...((((.-.....(((((((((..-...((((.....))))))))))))).......))))........))))).......-..--- ( -18.58) >consensus GGCCCAGAGAGAA____A_AUUCCGGU_UAAUCAUAGGCUAA__AACUGGUAAUCAAUACCUUAGCCUAUUUCAUGCCCGGAAAACAUAUUUC_UUUUUGU_UU___ ......................((((.......(((((((((......((((.....))))))))))))).......)))).......................... (-12.81 = -12.92 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:41 2006