
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,789,423 – 13,789,579 |
| Length | 156 |
| Max. P | 0.999388 |

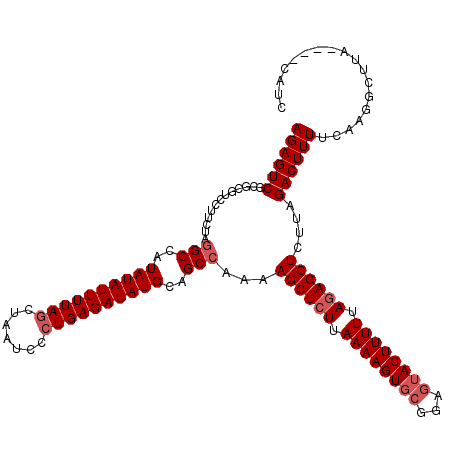
| Location | 13,789,423 – 13,789,539 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -28.60 |
| Energy contribution | -29.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13789423 116 + 23771897 AGAGUCGCGCGUCCUUCUAGGCCAUAUAUUUUAGCUAAUCCCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUUCUAGACUUUUCAAGGCUUA----CAUC .(((((.........(((((((..((((((((((.......))))))))))..)))..(((((((.((((((((...)))))))).))))))).))))........))))).----.... ( -34.73) >DroSec_CAF1 26454 116 + 1 AGAGUCGCGCGUCCUUCUAGGCCAUAUAUUUUAGCUAAUCCCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUUCGGACUACUUUUUAGAGCUCUUAGACUUUUCAAGGCUUA----CAUC .(((((.........(((((((..((((((((((.......))))))))))..)))...((((((.((((((.......)))))).))))))..))))........))))).----.... ( -31.03) >DroSim_CAF1 22586 116 + 1 AGAGUCGCGCGUCCUUCUAUGCCAUAUAUUUUACCUAAUCUCUGAGAUAUGCAGCCCAAAGCUCUUAAAAGUGCGGAGUACUUUUUACAGCUCUUAGACUUUUCAAGGCUUA----CAUC ((((((((((((......))))..(((((((((.........)))))))))..))....((((...((((((((...))))))))...))))....))))))..........----.... ( -21.60) >DroEre_CAF1 25908 116 + 1 AGAGUCGCGCGUCCUUCUAGGCCAUAUAUUUUAGCUAAUCCCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUCUUAGACUUUUCAAGGCUUA----CAUC .(((((.........(((((((..((((((((((.......))))))))))..)))...((((((.((((((((...)))))))).))))))..))))........))))).----.... ( -33.93) >DroYak_CAF1 24803 120 + 1 AGAGUCGCGCUUCCUUGUAGGCCAUAUAUUUUAGCUAAUCCCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUCCUAGACUUUUCAGGGCUUACAUACAUC ...............(((((((..((((((((((.......))))))))))..)))..(((((((.((((((..(((((.((....)).)))))...)))))).)))))))...)))).. ( -35.80) >consensus AGAGUCGCGCGUCCUUCUAGGCCAUAUAUUUUAGCUAAUCCCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUCUUAGACUUUUCAAGGCUUA____CAUC ((((((.............(((..((((((((((.......))))))))))..)))...((((((.((((((((...)))))))).))))))....)))))).................. (-28.60 = -29.60 + 1.00)


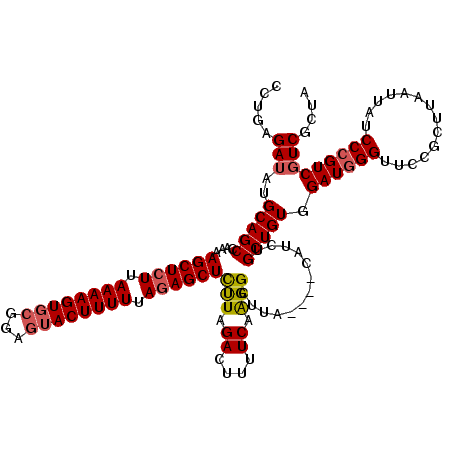
| Location | 13,789,463 – 13,789,579 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13789463 116 + 23771897 CCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUUCUAGACUUUUCAAGGCUUA----CAUCUGCUGUGGAUGGGUUCCGCUUAAUUAUCCCGUCGUCGCUA ............(((...(((((((.((((((((...)))))))).)))))))...(((....((.(((...----.....))).))((((((..............)))))))))))). ( -33.64) >DroSec_CAF1 26494 116 + 1 CCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUUCGGACUACUUUUUAGAGCUCUUAGACUUUUCAAGGCUUA----CAUCUGCUGUGGAUGGGUUCCGCUUAAUUAUCCCGUCGUCGCUA ............(((....((((((.((((((.......)))))).))))))....(((....((.(((...----.....))).))((((((..............)))))))))))). ( -29.84) >DroSim_CAF1 22626 116 + 1 UCUGAGAUAUGCAGCCCAAAGCUCUUAAAAGUGCGGAGUACUUUUUACAGCUCUUAGACUUUUCAAGGCUUA----CAUCUGCUGUGGAUAGGUUCCGCUUAAUUAUCCCGUCUUCGCUA ...(((((..((((....((((.(((((((((..(((((..........)))))...)))))).))))))).----...)))).(((((.....)))))...........)))))..... ( -27.80) >DroEre_CAF1 25948 116 + 1 CCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUCUUAGACUUUUCAAGGCUUA----CAUCUGCUGUGGAUGGGUUCCGCUUAAUUAUCCCGUCGUCGCUA ............(((....((((((.((((((((...)))))))).))))))....(((....((.(((...----.....))).))((((((..............)))))))))))). ( -32.74) >DroYak_CAF1 24843 120 + 1 CCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUCCUAGACUUUUCAGGGCUUACAUACAUCUGCUGUGGAUGGGCUCCGCUUAAUUAUCCCGUCGUCGCUA ...(.((((.((((((..(((((((.((((((..(((((.((....)).)))))...)))))).))))))).....(((((.....)))))))))..)).....)))).).......... ( -34.90) >consensus CCUGAGAUAUGCAGCCAAAAGCUCUUAAAAGUGCGGAGUACUUUUUAGAGCUCUUAGACUUUUCAAGGCUUA____CAUCUGCUGUGGAUGGGUUCCGCUUAAUUAUCCCGUCGUCGCUA .....(((..(((((....((((((.((((((((...)))))))).))))))(((.((....)).))).............))))).((((((..............))))))))).... (-27.34 = -27.78 + 0.44)


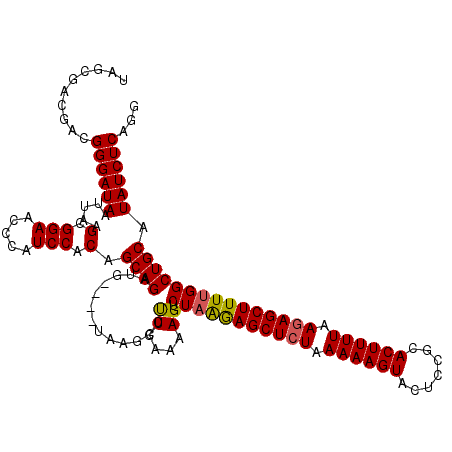
| Location | 13,789,463 – 13,789,579 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -31.04 |
| Energy contribution | -30.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13789463 116 - 23771897 UAGCGACGACGGGAUAAUUAAGCGGAACCCAUCCACAGCAGAUG----UAAGCCUUGAAAAGUCUAGAAGCUCUAAAAAGUACUCCGCACUUUUAAGAGCUUUUGGCUGCAUAUCUCAGG ..........((((((.(((((((((.....)))(((.....))----)..).)))))..((.(((((((((((.((((((.......)))))).)))))))))))))...))))))... ( -31.80) >DroSec_CAF1 26494 116 - 1 UAGCGACGACGGGAUAAUUAAGCGGAACCCAUCCACAGCAGAUG----UAAGCCUUGAAAAGUCUAAGAGCUCUAAAAAGUAGUCCGAACUUUUAAGAGCUUUUGGCUGCAUAUCUCAGG ..........((((((.(((((((((.....)))(((.....))----)..).)))))..((.(((((((((((.((((((.......)))))).)))))))))))))...))))))... ( -32.00) >DroSim_CAF1 22626 116 - 1 UAGCGAAGACGGGAUAAUUAAGCGGAACCUAUCCACAGCAGAUG----UAAGCCUUGAAAAGUCUAAGAGCUGUAAAAAGUACUCCGCACUUUUAAGAGCUUUGGGCUGCAUAUCUCAGA ..........((((((.....(.(((.....))).).((((...----.(((((((((((.(((..((.(((......))).))..).))))))))).))))....)))).))))))... ( -29.50) >DroEre_CAF1 25948 116 - 1 UAGCGACGACGGGAUAAUUAAGCGGAACCCAUCCACAGCAGAUG----UAAGCCUUGAAAAGUCUAAGAGCUCUAAAAAGUACUCCGCACUUUUAAGAGCUUUUGGCUGCAUAUCUCAGG ..........((((((.(((((((((.....)))(((.....))----)..).)))))..((.(((((((((((.((((((.......)))))).)))))))))))))...))))))... ( -31.80) >DroYak_CAF1 24843 120 - 1 UAGCGACGACGGGAUAAUUAAGCGGAGCCCAUCCACAGCAGAUGUAUGUAAGCCCUGAAAAGUCUAGGAGCUCUAAAAAGUACUCCGCACUUUUAAGAGCUUUUGGCUGCAUAUCUCAGG ..((...((.(((....((.....)).))).))....)).((.(((((((.(((((....))...(((((((((.((((((.......)))))).))))))))))))))))))).))... ( -34.20) >consensus UAGCGACGACGGGAUAAUUAAGCGGAACCCAUCCACAGCAGAUG____UAAGCCUUGAAAAGUCUAAGAGCUCUAAAAAGUACUCCGCACUUUUAAGAGCUUUUGGCUGCAUAUCUCAGG ..........((((((.....(.(((.....))).).((((.............((....)).(((((((((((.((((((.......)))))).))))))))))))))).))))))... (-31.04 = -30.88 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:01 2006