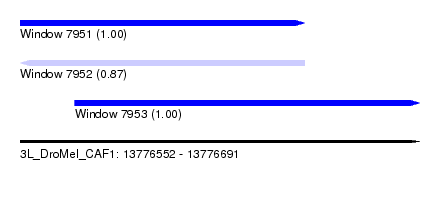
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,776,552 – 13,776,691 |
| Length | 139 |
| Max. P | 0.999883 |

| Location | 13,776,552 – 13,776,651 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.78 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
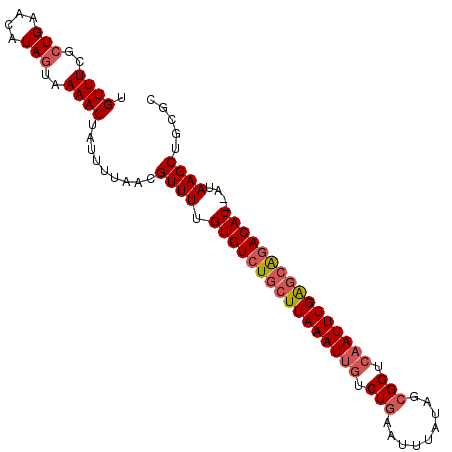
>3L_DroMel_CAF1 13776552 99 + 23771897 UGUUUCGCUGAACAUAGUAAAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCAGACAUAUAUAAGCCGCGC .....(((((((.((((.....))))))))..(((((((((((((((((((((.(((.........))).)))))))))))))))))...)))).))). ( -31.70) >DroSec_CAF1 23125 97 + 1 UGUUUCGCUGAACAUAGUAAAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCACACAU--AUAAGCAGCGC .....(((((...((((.....))))..........(((((.(((((((((((.(((.........))).))))))))))).))))--)....))))). ( -28.10) >DroSim_CAF1 18859 97 + 1 UGUUUCGCUGAACAUAGUAAAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCAGACAU--AUAAGCUGCGC .....(((((((.((((.....))))))))..(((((((((((((((((((((.(((.........))).))))))))))))))))--).)))).))). ( -33.10) >DroEre_CAF1 22614 97 + 1 UGUUUCGCUGAACAUAAUAAAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUAUUUUUAUUGCGGUCAAUUUGAGCAGACAU--AUAAGCUACGC (((((....)))))..................(((((((((((((((((((((.((...........)).))))))))))))))))--).))))..... ( -27.10) >DroYak_CAF1 21383 97 + 1 UGUUUCGCUGAGCAUAGUAAAACUAUUUUAACGUUUUGUGUCUGCUUAAAUCAUCUGCUUUUAUUGCGGUCAAUUUGGUGGGACAU--CAAAGCUACGC .((((..(((....)))..)))).........(((((((((((..((((((...((((.......))))...))))))..))))).--))))))..... ( -22.70) >consensus UGUUUCGCUGAACAUAGUAAAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCAGACAU__AUAAGCUGCGC .((((..(((....)))..)))).........((((.((((((((((((((((.(((.........))).))))))))))))))))....))))..... (-24.70 = -25.78 + 1.08)

| Location | 13,776,552 – 13,776,651 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.00 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.01 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13776552 99 - 23771897 GCGCGGCUUAUAUAUGUCUGCUCAAAUUGACCGCUAUAAAUUCAGACAAUUUAAGCAGACACAAAACGUUAAAAUAGUUUUACUAUGUUCAGCGAAACA .(((.....(((((((((((((.((((((....((........)).)))))).)))))))).(((((.........)))))..)))))...)))..... ( -22.00) >DroSec_CAF1 23125 97 - 1 GCGCUGCUUAU--AUGUGUGCUCAAAUUGACCGCUAUAAAUUCAGACAAUUUAAGCAGACACAAAACGUUAAAAUAGUUUUACUAUGUUCAGCGAAACA .(((((..(((--((((.((((.((((((....((........)).)))))).)))).))).(((((.........)))))..))))..)))))..... ( -22.10) >DroSim_CAF1 18859 97 - 1 GCGCAGCUUAU--AUGUCUGCUCAAAUUGACCGCUAUAAAUUCAGACAAUUUAAGCAGACACAAAACGUUAAAAUAGUUUUACUAUGUUCAGCGAAACA .(((.......--.((((((((.((((((....((........)).)))))).))))))))............((((.....)))).....)))..... ( -21.20) >DroEre_CAF1 22614 97 - 1 GCGUAGCUUAU--AUGUCUGCUCAAAUUGACCGCAAUAAAAAUAGACAAUUUAAGCAGACACAAAACGUUAAAAUAGUUUUAUUAUGUUCAGCGAAACA .(((.(..(((--(((((((((.((((((.................)))))).)))))))).(((((.........)))))..))))..).)))..... ( -19.53) >DroYak_CAF1 21383 97 - 1 GCGUAGCUUUG--AUGUCCCACCAAAUUGACCGCAAUAAAAGCAGAUGAUUUAAGCAGACACAAAACGUUAAAAUAGUUUUACUAUGCUCAGCGAAACA ((..(((((((--.((((.....((((((.(.((.......)).).)))))).....))))))))........((((.....)))))))..))...... ( -14.40) >consensus GCGCAGCUUAU__AUGUCUGCUCAAAUUGACCGCUAUAAAUUCAGACAAUUUAAGCAGACACAAAACGUUAAAAUAGUUUUACUAUGUUCAGCGAAACA .(((..........((((((((.((((((.................)))))).)))))))).(((((.........)))))..........)))..... (-15.57 = -16.01 + 0.44)

| Location | 13,776,571 – 13,776,691 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -25.34 |
| Energy contribution | -26.70 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
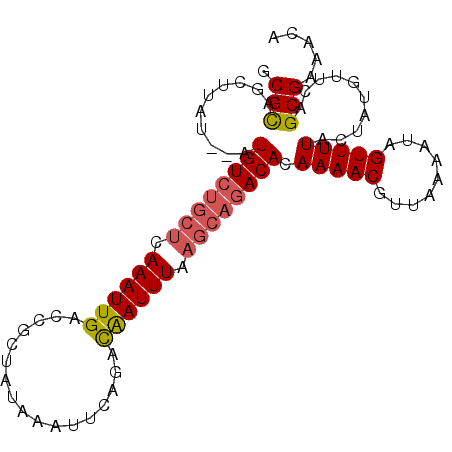
>3L_DroMel_CAF1 13776571 120 + 23771897 AAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCAGACAUAUAUAAGCCGCGCCAAAUUGCUUAGUUAGCUAAAUAAAAGAAAACUAAAUCUC .............(((((((((((((((((((((.(((.........))).)))))))))))))))))...)))).(((......)))((((((..((.......))..))))))..... ( -31.90) >DroSec_CAF1 23144 118 + 1 AAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCACACAU--AUAAGCAGCGCCAAAUUGCUUAGUUAGCUGAAGAAAAGAAAACUAAAUCUC .............(((((((((.(((((((((((.(((.........))).))))))))))).))))--).)))).(((......)))((((((..((.......))..))))))..... ( -28.70) >DroSim_CAF1 18878 118 + 1 AAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCAGACAU--AUAAGCUGCGCCAAAUUGCUUAGUUAGCUGAAGAAAAGAAAACUAAAUCUC .............(((((((((((((((((((((.(((.........))).))))))))))))))))--).)))).(((......)))((((((..((.......))..))))))..... ( -34.20) >DroEre_CAF1 22633 118 + 1 AAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUAUUUUUAUUGCGGUCAAUUUGAGCAGACAU--AUAAGCUACGCGAAAUUGCUUAGUUAGCUGAAGAUAGAAAAACUAGCACUC ...(((((((...(((.(((((((((((((((((.((...........)).))))))))))))))))--)..(((((.(((....))).))))))))..))))))).............. ( -34.50) >DroYak_CAF1 21402 118 + 1 AAACUAUUUUAACGUUUUGUGUCUGCUUAAAUCAUCUGCUUUUAUUGCGGUCAAUUUGGUGGGACAU--CAAAGCUACGCGCAAUGGCUCAGUUAGCCGAAGAAAAGAAAACUAGCACUG .............(((((((((((..((((((...((((.......))))...))))))..))))).--)))))).....((..(((((.....)))))......((....)).)).... ( -24.70) >consensus AAACUAUUUUAACGUUUUGUGUCUGCUUAAAUUGUCUGAAUUUAUAGCGGUCAAUUUGAGCAGACAU__AUAAGCUGCGCCAAAUUGCUUAGUUAGCUGAAGAAAAGAAAACUAAAUCUC .............(((((((((((((((((((((.(((.........))).)))))))))))))))).....(((((.((......)).))))).............)))))........ (-25.34 = -26.70 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:56 2006