
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,773,790 – 13,774,021 |
| Length | 231 |
| Max. P | 0.999939 |

| Location | 13,773,790 – 13,773,906 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.26 |
| Mean single sequence MFE | -38.49 |
| Consensus MFE | -34.44 |
| Energy contribution | -34.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.69 |
| SVM RNA-class probability | 0.999939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13773790 116 + 23771897 AUGGCAGGGGUCCCCUUCUCAGCCUUCAGAGGCCAUAAUAUCUUUGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGCCCCCCU----C .((((.(((((..........((((....))))............)))))))))((.(((..(((.(((((((............))))))).)))..)))))............----. ( -38.15) >DroSec_CAF1 20442 114 + 1 AUGGCAGGGGGCCCAUUCUCAGCCUUCAGAGGCCAUAAUAUCUUCGCC-CGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCACCACAAGACCCCCC----- .(((..((((((.......((((((((.(.(((.(........).)))-)....))).))))).(.(((((((............))))))).))))))).)))...........----- ( -37.60) >DroSim_CAF1 15787 115 + 1 AUGGCAGGGGGCCCAUUCUCAGCCUUCAGAGGCCAUAAUAUCUUCGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGACCCCCC----- .((((..(((((.........((((....))))............)))))))))((.(((..(((.(((((((............))))))).)))..)))))............----- ( -37.90) >DroEre_CAF1 19912 120 + 1 AUGGCAGGGGGCCCAUUCUCAGCCUUCAGAGGCCAUAAUAUCUUUGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGUCCCCCCACUUU .(((..(((((((((((((..((...((((((........))))))....)).)))))))..(((.(((((((............))))))).)))...........))))))))).... ( -39.70) >DroYak_CAF1 18569 120 + 1 AUGGCAGGGGGCCCAUUCCCAGCCUUCGGAGGCCAUAAUAUCUUCGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGUCCACCCCCCUC ......((((((.......((((((((((.(((.(........).)))...)).))).))))).(.(((((((............))))))).))))))).................... ( -39.10) >consensus AUGGCAGGGGGCCCAUUCUCAGCCUUCAGAGGCCAUAAUAUCUUCGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGACCCCCC_____ ......((((((.......((((((((.(.(((.(........).))).)....))).))))).(.(((((((............))))))).))))))).................... (-34.44 = -34.64 + 0.20)

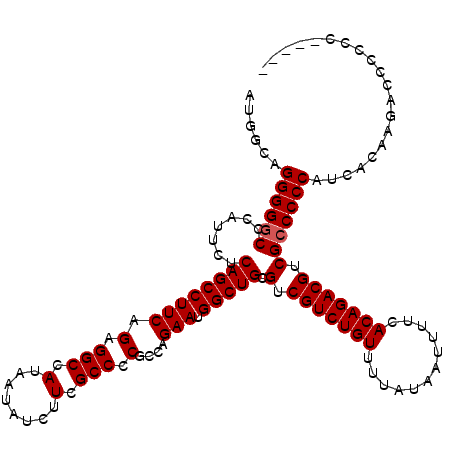
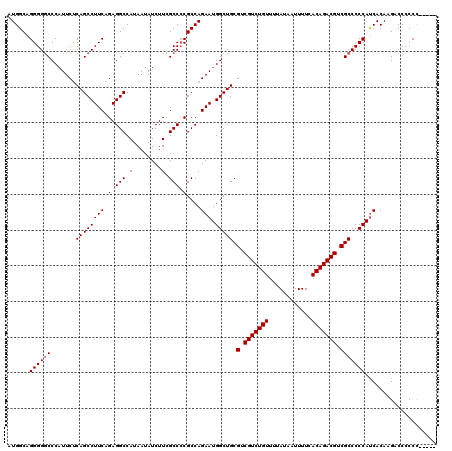
| Location | 13,773,790 – 13,773,906 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.26 |
| Mean single sequence MFE | -46.28 |
| Consensus MFE | -40.76 |
| Energy contribution | -41.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13773790 116 - 23771897 G----AGGGGGGCUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCAAAGAUAUUAUGGCCUCUGAAGGCUGAGAAGGGGACCCCUGCCAU .----((((((.((((((((((..(((.(((((((............))))))).)))..)))))...))))).)...........(((((....)))))..........)))))..... ( -42.70) >DroSec_CAF1 20442 114 - 1 -----GGGGGGUCUUGUGGUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCG-GGCGAAGAUAUUAUGGCCUCUGAAGGCUGAGAAUGGGCCCCCUGCCAU -----(((((((((..((((((((((..(((((((............))))))).(((.(((.....))).-.)))...........))))))....)))).....)))))))))..... ( -47.10) >DroSim_CAF1 15787 115 - 1 -----GGGGGGUCUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCGAAGAUAUUAUGGCCUCUGAAGGCUGAGAAUGGGCCCCCUGCCAU -----(((((((((((((((((..(((.(((((((............))))))).)))..)))))...))))))).....((((..(((((....)))))...))))..)))))...... ( -46.70) >DroEre_CAF1 19912 120 - 1 AAAGUGGGGGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCAAAGAUAUUAUGGCCUCUGAAGGCUGAGAAUGGGCCCCCUGCCAU ...(..((....))..)(((((..(((.(((((((............))))))).)))..))))).(((((((((.....((((..(((((....)))))...)))).)))))..)))). ( -48.50) >DroYak_CAF1 18569 120 - 1 GAGGGGGGUGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCGAAGAUAUUAUGGCCUCCGAAGGCUGGGAAUGGGCCCCCUGCCAU .((((((.(.(.((.((..(((((((..(((((((............))))))).(((.(((.....)))...)))...........)))))))...)).))...).).))))))..... ( -46.40) >consensus _____GGGGGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCGAAGAUAUUAUGGCCUCUGAAGGCUGAGAAUGGGCCCCCUGCCAU .................(((((..(((.(((((((............))))))).)))..))))).((((((((......((((..(((((....)))))...))))....)))))))). (-40.76 = -41.00 + 0.24)


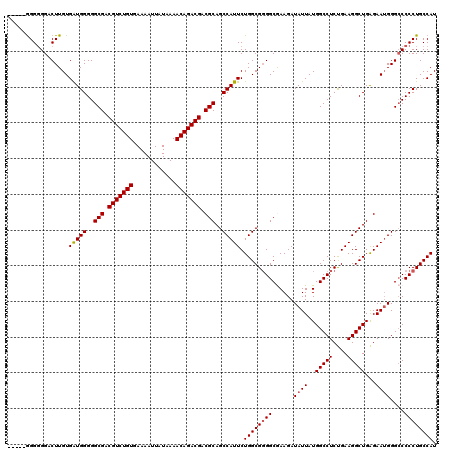
| Location | 13,773,830 – 13,773,946 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.59 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13773830 116 + 23771897 UCUUUGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGCCCCCCU----CCCCACCAUCUUUCAAGUGCCCUGGGUUGACACUCGCACAU ..........((.((.((((..(((.(((((((............))))))).)))..))))....(((((....----...(((..........)))....)))))....)).)).... ( -26.80) >DroSec_CAF1 20482 109 + 1 UCUUCGCC-CGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCACCACAAGACCCCCC----------CUACUUUCAAGUGCCCUGGGUUGACACUCGCACAU ........-.((.((..(((..(((.(((((((............))))))).)))..)))......(((((...----------.((((....))))....)))))....)).)).... ( -28.80) >DroSim_CAF1 15827 109 + 1 UCUUCGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGACCCCCC-----------CACUUUCAAGUGCCCUGGGUUGACACUCGCACAU ..........((.((.((((..(((.(((((((............))))))).)))..)))).....(((((...-----------((((....))))....)))))....)).)).... ( -30.90) >DroEre_CAF1 19952 118 + 1 UCUUUGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGUCCCCCCACUUUCCCCC--ACUUUCAAGUGCCCUGGGUUGACACUCGCACAA ..........((.((.((((..(((.(((((((............))))))).)))..)))).....(((..((((........(--(((....))))...))))..))).)).)).... ( -30.70) >DroYak_CAF1 18609 120 + 1 UCUUCGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGUCCACCCCCCUCCCCCCACACUUUCAAGUGCCCUGGGUUGACACUCGCACAA ..........((.((.((((..(((.(((((((............))))))).)))..)))).....(((.((((...........((((....))))....)))).))).)).)).... ( -31.26) >consensus UCUUCGCCCCGCCAGAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGACCCCCC_____CCC_C_CACUUUCAAGUGCCCUGGGUUGACACUCGCACAU ..........(((.....))).(((.(((((((............))))))).))).......................................((((...((((....)))))))).. (-25.20 = -25.20 + -0.00)



| Location | 13,773,830 – 13,773,946 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.59 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -35.00 |
| Energy contribution | -34.80 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13773830 116 - 23771897 AUGUGCGAGUGUCAACCCAGGGCACUUGAAAGAUGGUGGGG----AGGGGGGCUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCAAAGA .(((((((((.((..(((....((((........)))))))----...)).))))))(((((..(((.(((((((............))))))).)))..)))))........))).... ( -37.40) >DroSec_CAF1 20482 109 - 1 AUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUAG----------GGGGGGUCUUGUGGUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCG-GGCGAAGA .....((..((((((((((((((.((((....)))----------)....)))))).)))((..(((.(((((((............))))))).)))..))....)))))-..)).... ( -36.70) >DroSim_CAF1 15827 109 - 1 AUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUG-----------GGGGGGUCUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCGAAGA .....((((((((.......))))))))......-----------.....((((((((((((..(((.(((((((............))))))).)))..)))))...)))))))..... ( -38.80) >DroEre_CAF1 19952 118 - 1 UUGUGCGAGUGUCAACCCAGGGCACUUGAAAGU--GGGGGAAAGUGGGGGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCAAAGA ((((.((((((((.......))))))))...((--.((.....(..((....))..)(((((..(((.(((((((............))))))).)))..))))))).))...))))... ( -41.00) >DroYak_CAF1 18609 120 - 1 UUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUGUGGGGGGAGGGGGGUGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCGAAGA ((((.(.(.(.((..(((...(((((....)))))...)))..)).).).).((((((((((..(((.(((((((............))))))).)))..)))))...)))))))))... ( -39.70) >consensus AUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUG_G_GGG_____GGGGGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUCUGGCGGGGCGAAGA .(((.((((((((.......))))))))........................((((((((((..(((.(((((((............))))))).)))..)))))...)))))))).... (-35.00 = -34.80 + -0.20)

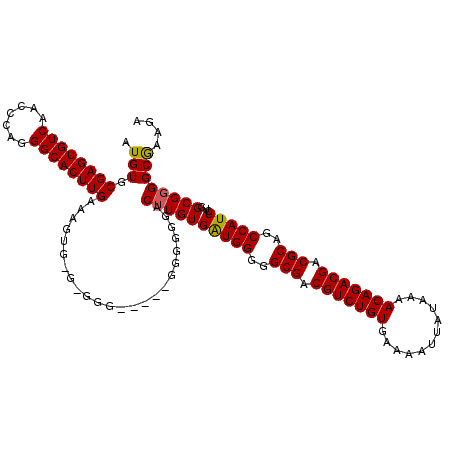

| Location | 13,773,906 – 13,774,021 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13773906 115 - 23771897 AUAAAUAAAAUAAAUGUAGGUGAAAAUAUACAUAAAUAAUGGGCACAAGCGCGGCUUUUUUUAUGAAGUGUUUGAAUGUGCGAGUGUCAACCCAGGGCACUUGAAAGAUGGUGGG .............(((((.((....)).)))))........(((((..((((((((((......))))).......)))))..)))))..((((...((.((....)))).)))) ( -22.91) >DroSec_CAF1 20556 110 - 1 AUAAAUAAAAUAAAUGUAGGUGAAAAUAUACAUAAAUAAUGGGCACAAGCGCGGCUUUUUUUAUGAAGUGUUUGAAUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUAG----- .............(((((.((....)).))))).........(((((..((..(((((......)))))...))..)))))(((((((.......)))))))........----- ( -22.60) >DroSim_CAF1 15902 109 - 1 AUAAAUAAAAUAAAUGUAGGUGAAAAUAUACAUAAAUAAUGGGCACAAGCGCGGCUUUUUUUAUGAAGUGUUUGAAUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUG------ .............(((((.((....)).))))).........(((((..((..(((((......)))))...))..)))))(((((((.......))))))).......------ ( -22.60) >DroEre_CAF1 20032 112 - 1 AUAAAUAAAAUAAAUGUAGGUGAAAAUAUACAUAAAUAAUGGGCACAAGCGCGGCUUU-UUUAUGAAGUGUUUGAUUGUGCGAGUGUCAACCCAGGGCACUUGAAAGU--GGGGG .............(((((.((....)).)))))........(((((..((((((((((-.....))))).......)))))..)))))..(((....((((....)))--).))) ( -25.01) >DroYak_CAF1 18689 115 - 1 AUAAAUAAAAUAAAUGUAGGUGAAAAUAUACAUAAAUAAUGGGCACAAGCGCGGCUUUUUUUAUGAAGUGUUUGAUUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUGUGGGGG .............(((((.((....)).)))))........(((((..((((((((((......))))).......)))))..)))))..(((...(((((....))))).))). ( -28.91) >consensus AUAAAUAAAAUAAAUGUAGGUGAAAAUAUACAUAAAUAAUGGGCACAAGCGCGGCUUUUUUUAUGAAGUGUUUGAAUGUGCGAGUGUCAACCCAGGGCACUUGAAAGUG_G_GGG .............(((((.((....)).))))).........((((((((((..(.........)..)))))))..))).((((((((.......))))))))............ (-22.10 = -22.10 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:51 2006