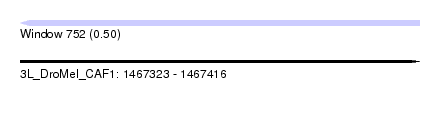
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,467,323 – 1,467,416 |
| Length | 93 |
| Max. P | 0.500000 |

| Location | 1,467,323 – 1,467,416 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
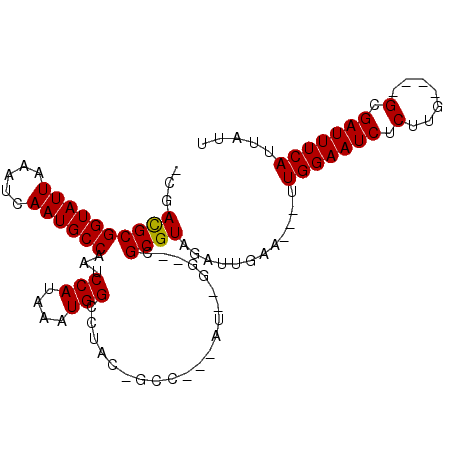
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1467323 93 - 23771897 -CGACGCGGUAUUAAAUCAAUGCCAAUCCAUAAAUGGCCUAC-GCCAUUAUGAGG--CGCGUAGAUUGAA---UUGGAAUCUCUUG----GCGAUUUCAUUAUU -..((((((((((.....))))))....(((((.((((....-)))))))))...--.))))........---.(((((((.....----..)))))))..... ( -24.80) >DroVir_CAF1 123278 89 - 1 UAGACGCGGUAUUAAAUCAAUGCCAAUCCAUAAAUGGUGCAU-------AC--GA--CGCGUCGAUUGUAUUAUUGGAAUCUCUUA----GCGAUUUCAUUAUU ....(((((((((.....))))))..((((...((((((((.-------.(--((--....)))..))))))))))))........----)))........... ( -22.70) >DroGri_CAF1 103749 96 - 1 UGGACGCGGUAUUAAAUCAAUGCCAAUCCAUAAAUGGGUCUCCGCA---GC--GAUGCGCGUAGAUUGAA---UUGGAAUCUCUCAGUGAGCGAUUUCAUUAUU ((((...((((((.....))))))..))))....(.((((((((((---..--..)))).).))))).).---.(((((((.(((...))).)))))))..... ( -25.70) >DroSec_CAF1 116023 93 - 1 -CGACGCGGUAUUAAAUCAAUGCCAAUCCAUAAAUGGCCUAC-GCCAUUAUGAGG--CGCGUAGAUAGAA---UUGGAAUCUCUUG----GCGAUUUCAUUAUU -..((((((((((.....))))))....(((((.((((....-)))))))))...--.))))........---.(((((((.....----..)))))))..... ( -24.80) >DroSim_CAF1 113137 93 - 1 -CGACGCGGUAUUAAAUCAAUGCCAAUCCAUAAAUGGCCUAC-GCCAUUAUGAGG--CGCGUAGAUUGAA---UUGGAAUCUCUUG----GCGAUUUCAUUAUU -..((((((((((.....))))))....(((((.((((....-)))))))))...--.))))........---.(((((((.....----..)))))))..... ( -24.80) >DroWil_CAF1 116058 84 - 1 --AAUGCGGUAUUAAAUCAAUGCCAAUCCAUAAAUGGCCUAC-GC--------GG--CGCGUAGAUUGAA---UUGGAAUCUCUUG----GCGAUUUCAUUAUU --.((((((((((.....))))))............(((...-..--------))--)))))........---.(((((((.....----..)))))))..... ( -18.90) >consensus _CGACGCGGUAUUAAAUCAAUGCCAAUCCAUAAAUGGCCUAC_GCC___AU__GG__CGCGUAGAUUGAA___UUGGAAUCUCUUG____GCGAUUUCAUUAUU ...((((((((((.....))))))...(((....))).....................))))............(((((((.(.......).)))))))..... (-16.74 = -16.60 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:39 2006