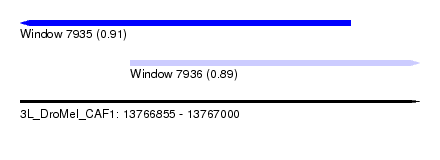
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,766,855 – 13,767,000 |
| Length | 145 |
| Max. P | 0.907893 |

| Location | 13,766,855 – 13,766,975 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -19.18 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
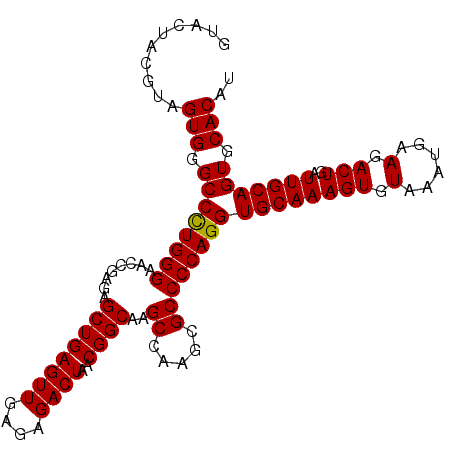
>3L_DroMel_CAF1 13766855 120 - 23771897 UACACUUUGCACCUGGGGCGCUUGGCUUGCCGUUAGUCUCUCAACUCAGCUCUCGGUUCCCAAGGCCCACUACGUAGUACCCACUACCAACUACCUACUACCUACUACCUACUACCUACU ...............((((.(((((...((((..((.((........))))..))))..))))))))).....(((((....)))))................................. ( -26.60) >DroSec_CAF1 13457 99 - 1 UACACUUUGCACCUGGGGCGCUUGGCUUGCCGUUAGUCUCUCAACUCAGC-------UCCCAGGGCCCACUACGUAGUACCCACUACCAACUACUCACUACCCACU-------------- ........((.(((((((.(((.((....))(((........)))..)))-------))))))))).......(((((....)))))...................-------------- ( -23.10) >DroSim_CAF1 8691 120 - 1 UACACUUUGCACCUGGGGCGCUUGGCUUGCCGUUAGUCUCUCAACUCAGCUCUCGGCUCCCAGGGCCCACUACGUAGUACCCACUACCAACUACUCACUACCCACUACCAACUACCUACU ...............((((.(((((...((((..((.((........))))..))))..))))))))).....(((((....)))))................................. ( -27.90) >DroEre_CAF1 12730 113 - 1 UACACUUUGCACCUGGGGCGCUUGGCUUGCCGUUAGUCUCCCAACUCAGCUCUUGGUUCCCAGGGCCCACUACCCACUACCCACUACCAAGGAUCCAGGACCCACU-------ACCCACU ...........((((((((.(((((...((((..((.((........))))..))))..)))))))))............((........))...)))).......-------....... ( -27.80) >DroYak_CAF1 10796 101 - 1 UACACUUUGCACCUGGGGCGCUUGGCUUGCCGUUAGUCUCUCAACUCAGCUCUCUGUUCCCAGGGCCCACUAGGCACU-------ACCCACUACCUUGGACCCCGG-------AC----- ............((((((.((((((...(((((((((......))).)))...(((....))))))...))))))...-------..(((......))).))))))-------..----- ( -27.90) >consensus UACACUUUGCACCUGGGGCGCUUGGCUUGCCGUUAGUCUCUCAACUCAGCUCUCGGUUCCCAGGGCCCACUACGUAGUACCCACUACCAACUACCCACUACCCACU_______ACC_ACU .......(((.....((((.(((((...((((..((.((........))))..))))..))))))))).....)))............................................ (-19.18 = -19.94 + 0.76)


| Location | 13,766,895 – 13,767,000 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.05 |
| Mean single sequence MFE | -33.58 |
| Consensus MFE | -28.48 |
| Energy contribution | -30.32 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13766895 105 + 23771897 GUACUACGUAGUGGGCCUUGGGAACCGAGAGCUGAGUUGAGAGACUAACGGCAAGCCAAGCGCCCCAGGUGCAAAGUGUAAAUGAAGACUGAUUGCAGUGCACAU (((((..(((((.((((((((...))))).((((((((....))))..))))..)))..(((((...)))))...................)))))))))).... ( -31.90) >DroSec_CAF1 13483 98 + 1 GUACUACGUAGUGGGCCCUGGGA-------GCUGAGUUGAGAGACUAACGGCAAGCCAAGCGCCCCAGGUGCAAAGUGUAAAUGAAGACUGAUUGCAGUGCACAU (((((..(((((..((((((((.-------(((.((((....))))...((....)).)))..)))))).))..(((.(......).))).)))))))))).... ( -33.50) >DroSim_CAF1 8731 105 + 1 GUACUACGUAGUGGGCCCUGGGAGCCGAGAGCUGAGUUGAGAGACUAACGGCAAGCCAAGCGCCCCAGGUGCAAAGUGUAAAUGAAGACUGAUUGCAGUGCACAU (((((..(((((..((((((((.((.....((((((((....))))..))))..((...)))))))))).))..(((.(......).))).)))))))))).... ( -34.50) >DroEre_CAF1 12763 105 + 1 GUAGUGGGUAGUGGGCCCUGGGAACCAAGAGCUGAGUUGGGAGACUAACGGCAAGCCAAGCGCCCCAGGUGCAAAGUGUAAAUGAAGACUGAUUGCAGUGCACAU ..........(((.((((((((......(.(((..(((((....)))))((....)).))).)))))))((((((((.(......).)))..))))))).))).. ( -33.20) >DroYak_CAF1 10819 103 + 1 --AGUGCCUAGUGGGCCCUGGGAACAGAGAGCUGAGUUGAGAGACUAACGGCAAGCCAAGCGCCCCAGGUGCAAAGUGUAAAUGAAGACUGAUUGCAGUGCACAU --.(..(((.(.((((.(((....)))...(((.((((....))))...((....)).)))))))))))..)...((((........((((....)))))))).. ( -34.80) >consensus GUACUACGUAGUGGGCCCUGGGAACCGAGAGCUGAGUUGAGAGACUAACGGCAAGCCAAGCGCCCCAGGUGCAAAGUGUAAAUGAAGACUGAUUGCAGUGCACAU ..........(((.((((((((........((((((((....))))..))))..((.....))))))))((((((((.(......).)))..))))))).))).. (-28.48 = -30.32 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:40 2006