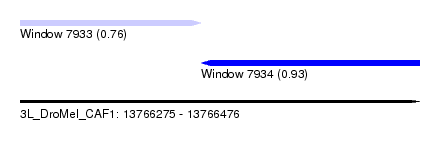
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,766,275 – 13,766,476 |
| Length | 201 |
| Max. P | 0.926999 |

| Location | 13,766,275 – 13,766,366 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -24.81 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13766275 91 + 23771897 GGUUGCCUCACCCUUAACAUCUGACUUGUAUUUGGCGUGAGUUGGUCG------GUUGUUGUUCGUUACAUGAAAGGGCAACUUUGUAACAACAGCA (..(..(((((((...(((.......)))....)).)))))..)..).------..(((((((.((((((....(((....)))))))))))))))) ( -26.30) >DroSec_CAF1 12877 91 + 1 GGUUGCCUCACCCUUAACAUCUGACUUGUAUUCGGCGUGAGUUGGUCG------GUUGUUGUUCGUUACAUGAAAGGGCAACUUUGUAACAACAGCA (..(..(((((((...(((.......)))....)).)))))..)..).------..(((((((.((((((....(((....)))))))))))))))) ( -25.60) >DroEre_CAF1 12136 97 + 1 GGUUCCCUCACCCUUAACAUCUGACUUGUAUUUGGCAUGAGUUGGUCGGGCUGGGUUGUUGUUCGUUACAUGAAAGGGCAACUUUGUAACAACAGCA .........((((....(..(..(((..(.......)..)))..)..)....))))(((((((.((((((....(((....)))))))))))))))) ( -27.40) >DroYak_CAF1 10161 91 + 1 GGCUCCCUCACCCUUAACAUCUGACUUGUGUUUGGCGUGAGUUGGUCG------CUUGUUGUUCGUUACAUGAAAGGGCAACUUUGUAACAACAGCA ((((..(((((((..(((((.......))))).)).)))))..)))).------..(((((((.((((((....(((....)))))))))))))))) ( -32.00) >consensus GGUUCCCUCACCCUUAACAUCUGACUUGUAUUUGGCGUGAGUUGGUCG______GUUGUUGUUCGUUACAUGAAAGGGCAACUUUGUAACAACAGCA ((((..(((((((...(((.......)))....)).)))))..)))).........(((((((.((((((....(((....)))))))))))))))) (-24.81 = -24.88 + 0.06)


| Location | 13,766,366 – 13,766,476 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 96.38 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -26.06 |
| Energy contribution | -27.46 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13766366 110 - 23771897 GGCUAACAAGUUGGGCAAGGGGUAGCUUAAGCGUCUAUCAACACGGUUUGUUGGAUUGCUUACUGGUUUUUAUUAUAAAACUGUGUACUUUGUU-UUGUUGUAUUUGGCCG (((((((((...((((((((((((((......).))))).(((((((((....((..(((....)))..))......))))))))).)))))))-)..))))...))))). ( -31.60) >DroSec_CAF1 12968 110 - 1 GGCUAACAAGUUGGGCAAGGGGUAGCUUAACCGUCUAUCAACACGGUUUGUUGGAUUGCUUACUGGUUUUUAUUAUAAAACUGUGUACUUUGUU-UUGUUGUAUUUGGCCG (((((((((...((((((((((((((......).))))).(((((((((....((..(((....)))..))......))))))))).)))))))-)..))))...))))). ( -31.60) >DroSim_CAF1 8202 110 - 1 GGCUAACAAGUUGGGCAAGGGGUAGCUUAACCGUCUAUCAACACGGUUUGUUGGAUUGCUUACUGGUUUUUAUUAUAAAACUGUGUACUUUGUU-UUGUUGUAUUUGGCCG (((((((((...((((((((((((((......).))))).(((((((((....((..(((....)))..))......))))))))).)))))))-)..))))...))))). ( -31.60) >DroEre_CAF1 12233 110 - 1 GGCAAACAAGCUGGGCAAGGGGUAGCUUAACCGUCUAUCAACACGGCUUGUUGGAUUGCUUACUGGUUUUUAUUAUAAAACUGUGUACACUGUU-UUGUUGUAUUUGGCCG (((((((((((((.......((((((......).)))))....))))))))....)))))....((((..(((.(((((((.((....)).)))-)))).)))...)))). ( -29.10) >DroYak_CAF1 10252 111 - 1 GGCUAACAAGUUGGCCAAGGGGUAGCUUAACCGUCUAUCAACACGGUUUGUUGCAUUGCUUACUGGUUUUUAUUAUAAAACUGUGUACUUUAUUUUUGUUGUAUUUGGCCG ((((((..(((.(((.((...(((((..((((((........)))))).))))).))))).)))((((((......))))))..((((..((....))..)))))))))). ( -28.20) >consensus GGCUAACAAGUUGGGCAAGGGGUAGCUUAACCGUCUAUCAACACGGUUUGUUGGAUUGCUUACUGGUUUUUAUUAUAAAACUGUGUACUUUGUU_UUGUUGUAUUUGGCCG (((((((((....(((((((((((((......).))))).(((((((((....((..(((....)))..))......))))))))).)))))))....))))...))))). (-26.06 = -27.46 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:38 2006