
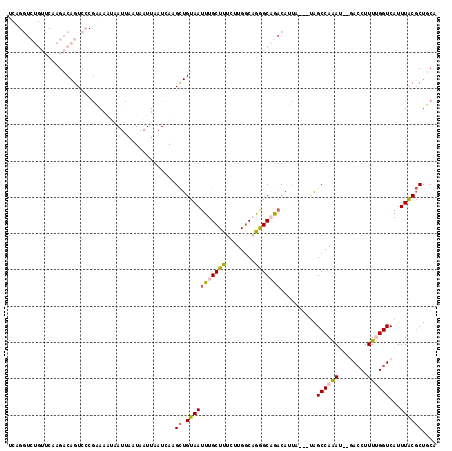
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,765,278 – 13,765,450 |
| Length | 172 |
| Max. P | 0.729855 |

| Location | 13,765,278 – 13,765,391 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -10.29 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13765278 113 + 23771897 UCAGGUCUGUUCAAGACAGUCCCGAAAAUAAUUUAUUAUUAAUCAUGCUGUAAUUUGCUUUCUUGGCAGGGCAGACAUUA---UGGCCAAAU--GACCUUUUGGUCAUUUACGCUUCA ....((((((((...(((((......(((((....)))))......)))))....((((.....))))))))))))....---.(((.((((--((((....))))))))..)))... ( -31.50) >DroVir_CAF1 8028 96 + 1 -------------GGUCAGUCC-GCA----AUCAUUAAUUCAUAAAGCUGUAAUUUGCUUU--UGGCCAAGCCAACAGCCGAGCAGCCAGAC--AACCUUUCGGUCAUUUGCGCUGCA -------------......((.-((.----.............(((((........)))))--((((...))))...)).))(((((((((.--.(((....)))..)))).))))). ( -25.90) >DroSec_CAF1 11898 113 + 1 UCAGGUCUGUUCAAGACAGUCCCAAAAAUAAUUAAUAAUUAAUCAUGCUGUAAUUUGCUUUCUUGGCAGGGCAGACAUUA---UGGCCAAAU--GACCUUUUGGUCAUUUACGCUUCA ....((((((((...(((((......(((........)))......)))))....((((.....))))))))))))....---.(((.((((--((((....))))))))..)))... ( -29.50) >DroEre_CAF1 11143 113 + 1 UCAGGUCUGUUCAAGACAGUCCCGAAAAUAAUUAAUAAUUAAUCAUGCUGUAAUUUGCUUUCUUGGCAGGGCAGACAUUA---UAGCCAAAU--GACCUUUUGGUCAUUUACGCUUCA ((.((.(((((...))))).)).)).....................(((((((((((((((......))))))))..)))---)))).((((--((((....))))))))........ ( -30.00) >DroYak_CAF1 9159 113 + 1 UCAGGUCUGUUCAAGACAGUCCCGAAAAUAAUUAAUAAUUAAUCAAGUUGUAAUUUGCUUUCUUGGCAGGGCAGACAUUA---UAGCCAAAU--GACCUUUUGGUCAUUUACUCCGCA ((.((.(((((...))))).)).)).....................(((((((((((((((......))))))))..)))---)))).((((--((((....))))))))........ ( -27.80) >DroMoj_CAF1 33955 94 + 1 -------------GGUCAGUCC-GCAA---AUCAUUAAUUCAUAAAGCUGUAAUUUGCUUU--UGACCGAGCCG--AGCCA---AGCCAGACAUAAGCUUUCGGUCAUUUGCGCUGCA -------------.(.(((..(-((((---(............(((((........)))))--(((((((...(--(((..---............))))))))))))))))))))). ( -23.54) >consensus UCAGGUCUGUUCAAGACAGUCCCGAAAAUAAUUAAUAAUUAAUCAAGCUGUAAUUUGCUUUCUUGGCAGGGCAGACAUUA___UAGCCAAAU__GACCUUUUGGUCAUUUACGCUGCA ..............................................((.(((((((((((........)))))))..........((((((........))))))...)))))).... (-10.29 = -10.35 + 0.06)


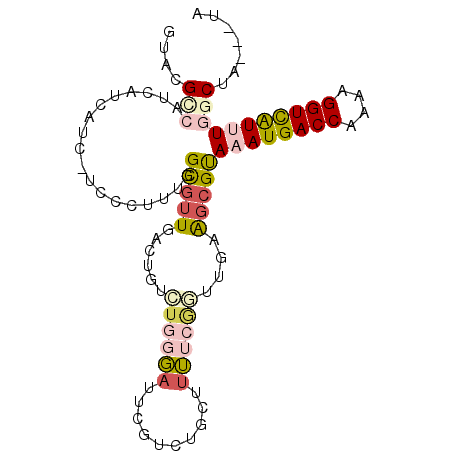
| Location | 13,765,356 – 13,765,450 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 70.94 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -12.35 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13765356 94 - 23771897 GUACGUCAUCAUCAUCUUCCCUUUGGCGUUGACUGUCUGGGAUUCGUCUGCUUUUCGCUUGAAGCGUAAAUGACCAAAAGGUCAUUUGGCCA---UA ....(((..........((((...((((.....)))).)))).......(((((......)))))..((((((((....)))))))))))..---.. ( -23.90) >DroVir_CAF1 8086 77 - 1 GUGUG-----------------CAUGUG-UG--UGUGGACUAUUUGUCUGCUUAUAAUUUGCAGCGCAAAUGACCGAAAGGUUGUCUGGCUGCUCGG .....-----------------....((-((--.(..(((.....)))..).))))....(((((.((.(..(((....)))..).))))))).... ( -27.90) >DroGri_CAF1 30427 78 - 1 GUGUG-----------------UUUGUGUUG--UGUGUACUAUUUGUCUGCUUAUAAAUUGCAGCGCAAAUGACCGAAAGGUUGUCUGCCUGCUCGA ....(-----------------((((((...--.((..((.....))..)).))))))).((((.(((.(..(((....)))..).))))))).... ( -22.70) >DroSec_CAF1 11976 94 - 1 GUACGCCAUCAUCAUCUUCCCUUUGGCGUUGACUGUCUGGGAUUCGUCUGCUUUUCGGUUGAAGCGUAAAUGACCAAAAGGUCAUUUGGCCA---UA ....(((..........((((...((((.....)))).)))).......(((((......)))))..((((((((....)))))))))))..---.. ( -26.60) >DroEre_CAF1 11221 94 - 1 GUACGCCAUCAUCAUCAUCCCUUUGGCGUUGACUGUCUGGGAUUCGUCUGCUUUUCGGUUGAAGCGUAAAUGACCAAAAGGUCAUUUGGCUA---UA ....(((.........(((((...((((.....)))).)))))......(((((......)))))..((((((((....)))))))))))..---.. ( -27.70) >DroYak_CAF1 9237 94 - 1 GUUCGCCAUCAUCAUCAUCCCUUUGGCGUUGACUGUCUGGGUUUCGUCUGCUUUUCGGUUGCGGAGUAAAUGACCAAAAGGUCAUUUGGCUA---UA ....(((...........(((...((((.....)))).))).....(((((.........)))))..((((((((....)))))))))))..---.. ( -25.60) >consensus GUACGCCAUCAUCAUC_UCCCUUUGGCGUUGACUGUCUGGGAUUCGUCUGCUUUUCGGUUGAAGCGUAAAUGACCAAAAGGUCAUUUGGCUA___UA ....(((..................(((((......((((((..........))))))....)))))((((((((....)))))))))))....... (-12.35 = -13.18 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:36 2006