| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,419,338 – 1,419,458 |
| Length | 120 |
| Max. P | 0.534707 |

| Location | 1,419,338 – 1,419,458 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.34 |
| Mean single sequence MFE | -35.97 |
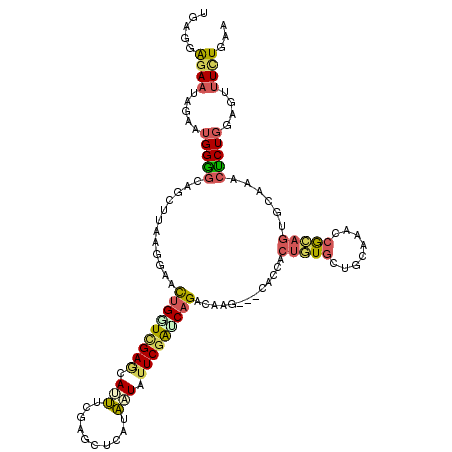
| Consensus MFE | -12.87 |
| Energy contribution | -12.68 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
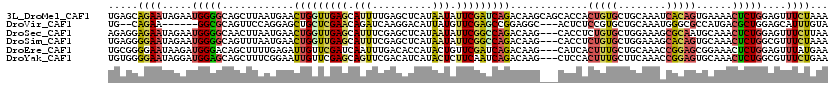
>3L_DroMel_CAF1 1419338 120 - 23771897 UGAGCAGAAUAGAAUGGGGCAGCUUAAUGAACUGGUUGAGCAUUUUGAGCUCAUAAUAUUCGAUCAGACAAGCAGCACCACUGUGCUGCAAAUCACAGUGAAAACUCUGGAGUUUCUAAA (((...(((((..(((((.(((((((((......)))))))....))..)))))..)))))..)))(((..(((((((....)))))))...(((.(((....))).))).)))...... ( -34.40) >DroVir_CAF1 51105 109 - 1 UG--CAGAA------GGCGCAGUUCCAGGAGCUGCUCGAACAGAUCAAGGACAUUAUGUUCGAGCCGGAGGC---ACUCUCCGUGCUGCAAAUGGGCGCCAUGACGCUGGAGCAUUUGUA ((--((((.------(((((....(((.((((.(((((((((..............)))))))))(((((..---...))))).))).)...)))))))).....((....)).)))))) ( -39.84) >DroSec_CAF1 69010 117 - 1 AGAGGAGAAUAGAAUGGGGCAACUUAAUGAACUGGUUGAGCAUUUCGAGCUCAUAAUAUUCGGCCAGACAAG---CACCUCUGUGCUGGAAAGCGCAAUGCAAACUCUGGAGUUUCUUAA .((((((..((((.((((....)))).....(((((((((.(((..(....)..))).)))))))))....(---((....((((((....)))))).)))....))))...)))))).. ( -33.50) >DroSim_CAF1 64964 117 - 1 UGAGGGGAAUAGAAUGGGGCAGUUUAAUGAACUGGUUGAGCAUUUCGAGCUCAUAAUAUUCGGCCAGACAAG---CACCUCUGUGCUGGAAAGCACAGUGCAAACUCUGGCGUUUCUAAA ....((((((.(((((...((((((...))))))..(((((.......)))))...))))).((((((...(---(((...((((((....))))))))))....))))))))))))... ( -41.60) >DroEre_CAF1 70735 117 - 1 UGCGGGGAAUAAGAUGGGACAGCUUUUGAGAUUGUUCGAUCAAUUUGACACCAUACUGUUCGAUCAGACAAG---CAUCACUUUGCUGCAAACCGGAGCGGAAACUCUGGAGUUUAUGAA (((((.(((...((((((((((((....)).))))))((((.....((((......)))).)))).......---))))..))).)))))..((((((......)))))).......... ( -29.30) >DroYak_CAF1 72930 117 - 1 UGUGGGGAAUAGGAUGGAGCAGCUUUCGGAAUUGUUCGAGCAGUUCGACAUCAUACUCUUCAAUCAGACAAG---CUCCACUUUGCUUCAAACCGGAGUGCAAACUCUGGCGUUUCUGAA ..((((((.((.((((..(.((((((((((....)))))).)))))..)))).)).)))))).(((((.(((---(........))))....(((((((....)))))))....))))). ( -37.20) >consensus UGAGGAGAAUAGAAUGGGGCAGCUUAAGGAACUGGUCGAGCAUUUCGAGCUCAUAAUAUUCGAUCAGACAAG___CACCACUGUGCUGCAAACCGCAGUGCAAACUCUGGAGUUUCUGAA .....((((.....(((((............(((((((((.(((..........))).))))))))).............(((((........)))))......)))))....))))... (-12.87 = -12.68 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:38 2006