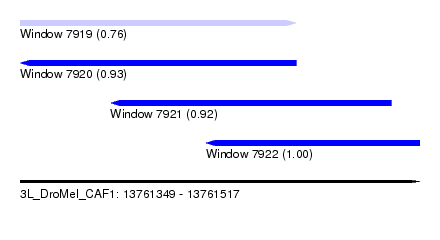
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,761,349 – 13,761,517 |
| Length | 168 |
| Max. P | 0.999835 |

| Location | 13,761,349 – 13,761,465 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -23.67 |
| Energy contribution | -25.28 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13761349 116 + 23771897 UUUAGUUAGACU--CUCAGUAAGUUUGACUGCGCAACUGUGUGGCAUUUCCGCUUCCGCAACGGAAGCUAAUGGAAAUGGAAGGGAGCAAUUGCGAGG--GUCCAAAUGACUUGGAAAUU ...(((((((((--(((.((((((((..(..(((....)))..)((((((((((((((...)))))))....))))))).....))))..))))))))--)))....)))))........ ( -37.90) >DroGri_CAF1 14621 108 + 1 UUUA---GGACUCAGUGAGUAUGUCUGACUGCGCGACUGUGUAGCACUUCCGCUUCCGUAACGGAAGCUAAUGAACGUAGU--CGAGUAGCAGCGACGCGAACAAAAUGGC-----AG-- ....---..((((...)))).((.(((.((((.(((((((((..((.....(((((((...)))))))...)).)))))))--)).)))))))))..((.(......).))-----..-- ( -30.90) >DroSec_CAF1 7887 116 + 1 UUUAGUUAGACU--CUGAGUAAGUUUGACUGCGCAACUGUGUGGCAUUUCCGCUUCCGCAACGGAAGCUAAUGGAAAUGGAAGGGAGCAAUUGCGAGG--GUCCAAAUGACUUGGAAAUU ...........(--(..(((...((.((((.(((((.(((....((((((((((((((...)))))))....))))))).......))).)))))..)--))).))...)))..)).... ( -38.20) >DroEre_CAF1 7145 116 + 1 UUUAGUUAGACU--CUCAGUAAGUUUGACUGCGCAACUGUGUGGCAUUUCCGCUUCCGCAACGGAAGCUAAUGGAAAUGGAAGGGAGCAAUUGCGAGG--GUCCAAAUGACUUGGAAAUU ...(((((((((--(((.((((((((..(..(((....)))..)((((((((((((((...)))))))....))))))).....))))..))))))))--)))....)))))........ ( -37.90) >DroYak_CAF1 5158 116 + 1 UUUAGUUAGACU--CUCAGUAAGUUUGACUGCGCAACUGUGUGGCAUUUCCGCUUCCGUAACGGAAGCUAAUGGAAAUGGAAGGGAACAAUUGCGAGG--GUCCAAAUGACUUGGAAAUU ...(((((((((--(((.((((((((..(..(((....)))..)((((((((((((((...)))))))....))))))).....))))..))))))))--)))....)))))........ ( -37.90) >DroMoj_CAF1 17698 108 + 1 UGUG---GGGCUCACUGAGUGUGUCUGACUGCGCGACUGUGUAGCACUUCCGCUUCCGCAACGGAAGCUAAUGAACAUAGC--CGAGCAGCAGCGACGCGAACAAAAUGGC-----AA-- ....---..(((...((..((((((((.((((.((.((((((..((.....(((((((...)))))))...)).)))))).--)).))))))..))))))..))....)))-----..-- ( -35.60) >consensus UUUAGUUAGACU__CUCAGUAAGUUUGACUGCGCAACUGUGUGGCAUUUCCGCUUCCGCAACGGAAGCUAAUGGAAAUGGAAGGGAGCAAUUGCGAGG__GUCCAAAUGACUUGGAAAUU ..(((((((((...........)))))))))(((((.(((....((((((((((((((...)))))))....))))))).......))).)))))......................... (-23.67 = -25.28 + 1.62)

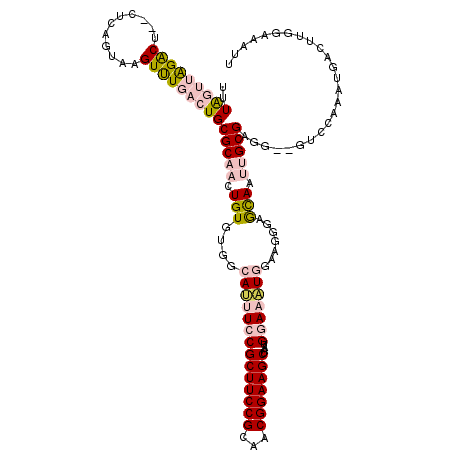
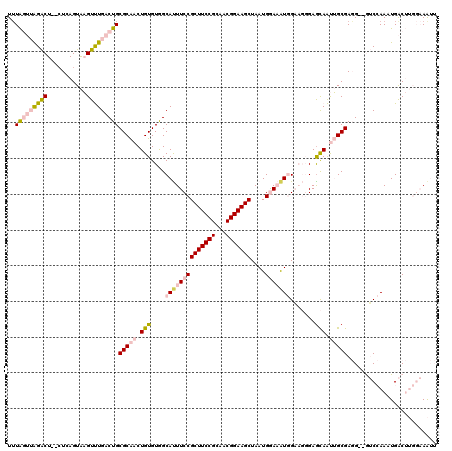
| Location | 13,761,349 – 13,761,465 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.56 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -23.09 |
| Energy contribution | -22.79 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13761349 116 - 23771897 AAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCACACAGUUGCGCAGUCAAACUUACUGAG--AGUCUAACUAAA ..............((((((--.((((((((((........(((((((....(((((((...)))))))))))))).....)))))))..(((.......))))))--.))))))..... ( -33.12) >DroGri_CAF1 14621 108 - 1 --CU-----GCCAUUUUGUUCGCGUCGCUGCUACUCG--ACUACGUUCAUUAGCUUCCGUUACGGAAGCGGAAGUGCUACACAGUCGCGCAGUCAGACAUACUCACUGAGUCC---UAAA --..-----..............((((((((....((--(((..((.((((.(((((((...)))))))...))))..))..))))).)))))..)))..((((...))))..---.... ( -28.30) >DroSec_CAF1 7887 116 - 1 AAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCACACAGUUGCGCAGUCAAACUUACUCAG--AGUCUAACUAAA ...(((..(((......(((--...((((((((........(((((((....(((((((...)))))))))))))).....))))))))..)))......)))..)--)).......... ( -30.62) >DroEre_CAF1 7145 116 - 1 AAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCACACAGUUGCGCAGUCAAACUUACUGAG--AGUCUAACUAAA ..............((((((--.((((((((((........(((((((....(((((((...)))))))))))))).....)))))))..(((.......))))))--.))))))..... ( -33.12) >DroYak_CAF1 5158 116 - 1 AAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGUUCCCUUCCAUUUCCAUUAGCUUCCGUUACGGAAGCGGAAAUGCCACACAGUUGCGCAGUCAAACUUACUGAG--AGUCUAACUAAA ..............((((((--.(((((((((((.......(((((((....(((((((...))))))))))))))....))))))))..(((.......))))))--.))))))..... ( -34.80) >DroMoj_CAF1 17698 108 - 1 --UU-----GCCAUUUUGUUCGCGUCGCUGCUGCUCG--GCUAUGUUCAUUAGCUUCCGUUGCGGAAGCGGAAGUGCUACACAGUCGCGCAGUCAGACACACUCAGUGAGCCC---CACA --..-----((((((..((..(.(((...(((((.((--(((.(((......(((((((((.....)))))))))...))).))))).)))))..))).)))..)))).))..---.... ( -38.80) >consensus AAUUUCCAAGUCAUUUGGAC__CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCACACAGUUGCGCAGUCAAACUUACUCAG__AGUCUAACUAAA .................(((.....((((((((........(((((((....(((((((...)))))))))))))).....))))))))..))).......................... (-23.09 = -22.79 + -0.30)
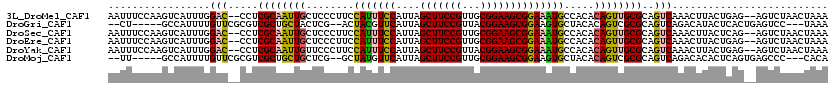


| Location | 13,761,387 – 13,761,505 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13761387 118 - 23771897 UUAAAAAUUGCUUGUACAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCAC .....((((((......((((.....))))((((((((((.....))))))....)))).--....)))))).........(((((((....(((((((...)))))))))))))).... ( -33.10) >DroSec_CAF1 7925 118 - 1 UUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCAC .........((...(((((((.....))))((((((((((.....))))))....)))).--....)))...)).......(((((((....(((((((...)))))))))))))).... ( -33.90) >DroSim_CAF1 4505 118 - 1 UUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCAC .........((...(((((((.....))))((((((((((.....))))))....)))).--....)))...)).......(((((((....(((((((...)))))))))))))).... ( -33.90) >DroEre_CAF1 7183 118 - 1 UUAAAAAUUGCUUCUACAUUGUUUUUCAAUUCCAGCUCGGAAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCAC .....((((((................((((((.....))))))((((((...)))))).--....)))))).........(((((((....(((((((...)))))))))))))).... ( -30.10) >DroYak_CAF1 5196 118 - 1 UUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGAC--CCUCGCAAUUGUUCCCUUCCAUUUCCAUUAGCUUCCGUUACGGAAGCGGAAAUGCCAC .....((((((......((((.....))))((((((((((.....))))))....)))).--....)))))).........(((((((....(((((((...)))))))))))))).... ( -33.10) >DroMoj_CAF1 17735 109 - 1 UUAACAUUUGCCUG-GCAUUUUCUUAGAGUCUCGAGUCG---UU-----GCCAUUUUGUUCGCGUCGCUGCUGCUCG--GCUAUGUUCAUUAGCUUCCGUUGCGGAAGCGGAAGUGCUAC ............((-((((((((....(((..(((((.(---(.-----((.....((....))..)).)).)))))--)))..........(((((((...))))))))))))))))). ( -31.30) >consensus UUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGAC__CCUCGCAAUUGCUCCCUUCCAUUUCCAUUAGCUUCCGUUGCGGAAGCGGAAAUGCCAC .....((((((......((((.....))))((((((((((.....))))))....)))).......)))))).........(((((((....(((((((...)))))))))))))).... (-26.19 = -26.82 + 0.63)

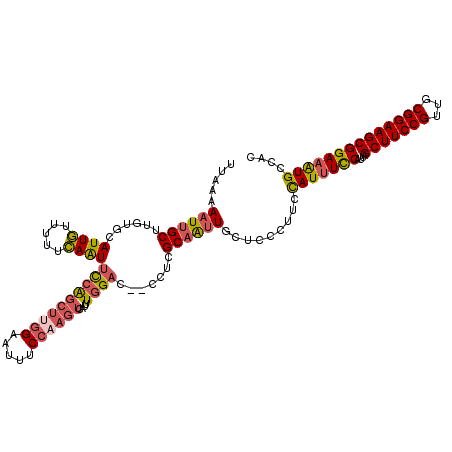
| Location | 13,761,427 – 13,761,517 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -16.22 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.20 |
| SVM RNA-class probability | 0.999835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13761427 90 - 23771897 CCCUUUAUUUAUUUAAAAAUUGCUUGUACAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGACCCUCGCAAUUGCUCCCUU .................((((((......((((.....))))((((((((((.....))))))....)))).....))))))........ ( -16.50) >DroSec_CAF1 7965 90 - 1 CCCUUUAUUUAUUUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGACCCUCGCAAUUGCUCCCUU .....................((...(((((((.....))))((((((((((.....))))))....)))).....)))...))...... ( -17.30) >DroSim_CAF1 4545 90 - 1 CCCUUUAUUUAUUUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGACCCUCGCAAUUGCUCCCUU .....................((...(((((((.....))))((((((((((.....))))))....)))).....)))...))...... ( -17.30) >DroEre_CAF1 7223 90 - 1 CCCUUUAUUUAUUUAAAAAUUGCUUCUACAUUGUUUUUCAAUUCCAGCUCGGAAUUUCCAAGUCAUUUGGACCCUCGCAAUUGCUCCCUU .................((((((................((((((.....))))))((((((...)))))).....))))))........ ( -13.50) >DroYak_CAF1 5236 90 - 1 CCCUUUAUUUAUUUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGACCCUCGCAAUUGUUCCCUU .................((((((......((((.....))))((((((((((.....))))))....)))).....))))))........ ( -16.50) >consensus CCCUUUAUUUAUUUAAAAAUUGCUUGUGCAUUGUUUUUCAAUUCCAGCUUGGAAUUUCCAAGUCAUUUGGACCCUCGCAAUUGCUCCCUU .................((((((......((((.....))))((((((((((.....))))))....)))).....))))))........ (-15.70 = -15.90 + 0.20)

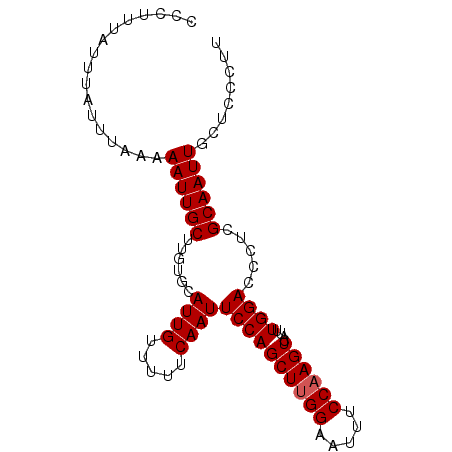

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:27 2006