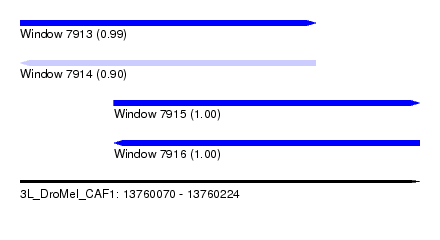
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,760,070 – 13,760,224 |
| Length | 154 |
| Max. P | 0.999867 |

| Location | 13,760,070 – 13,760,184 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -28.02 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13760070 114 + 23771897 AAUAAUUGAUGCCAGCUCUUGAAGAGGAAGCCCGUGGCUUGUGUUCUUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAG ...............(((.....)))((((((.((((((((((((.....(((..-....))).(((.((((((........)))))))))..))))))).))))).)))))).. ( -31.20) >DroSec_CAF1 6636 114 + 1 AAUAAUUGAUGCCAGCUCUUGAAGAGGAAGCCCGUGGCUUGUGUUCCUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAG ...............(((.....)))((((((.((((((((((((.....(((..-....))).(((.((((((........)))))))))..))))))).))))).)))))).. ( -31.20) >DroSim_CAF1 2769 114 + 1 AAUAAUUGAUGCCAGCUCUUGAAGAGGAAGCCCGUGGCUUGUGUUCUUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAG ...............(((.....)))((((((.((((((((((((.....(((..-....))).(((.((((((........)))))))))..))))))).))))).)))))).. ( -31.20) >DroEre_CAF1 5799 114 + 1 AAUAAUUGAUGCCAGGUCUUGAAGAGGAAGACCGUGGCUUGUGUUCUUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCCGGCUUCAG ......(((.(((.((((((.......))))))((((((((((((.....(((..-....))).(((.((((((........)))))))))..))))))).))))).))).))). ( -32.40) >DroYak_CAF1 2873 115 + 1 AAUAAUUGAUGCCAGUUCUUGAAGAGGAAGCCCGUGGCUUGUGUUCUUUCUGAUCCCUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAG ...(((((....))))).........((((((.(((((((((((((.....))...........(((.((((((........)))))))))...)))))).))))).)))))).. ( -29.80) >consensus AAUAAUUGAUGCCAGCUCUUGAAGAGGAAGCCCGUGGCUUGUGUUCUUUCUGAUC_CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAG ...............(((.....)))((((((.(((((((((((((.....))...........(((.((((((........)))))))))...)))))).))))).)))))).. (-28.02 = -28.30 + 0.28)



| Location | 13,760,070 – 13,760,184 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 97.99 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.04 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13760070 114 - 23771897 CUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAAGAACACAAGCCACGGGCUUCCUCUUCAAGAGCUGGCAUCAAUUAUU ..((((((((.(((......((((((((((((..........))))).))).))))...-..((.....))......))).))))))))(((.....)))............... ( -25.40) >DroSec_CAF1 6636 114 - 1 CUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAGGAACACAAGCCACGGGCUUCCUCUUCAAGAGCUGGCAUCAAUUAUU ..((((((((.(((......((((((((((((..........))))).))).))))..(-..((....))..)....))).))))))))(((.....)))............... ( -25.70) >DroSim_CAF1 2769 114 - 1 CUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAAGAACACAAGCCACGGGCUUCCUCUUCAAGAGCUGGCAUCAAUUAUU ..((((((((.(((......((((((((((((..........))))).))).))))...-..((.....))......))).))))))))(((.....)))............... ( -25.40) >DroEre_CAF1 5799 114 - 1 CUGAAGCCGGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAAGAACACAAGCCACGGUCUUCCUCUUCAAGACCUGGCAUCAAUUAUU ((((.(((...)))......((((((((((((..........))))).))).))))...-..))))...........((((.((((((.......)))))))))).......... ( -25.10) >DroYak_CAF1 2873 115 - 1 CUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAGGGAUCAGAAAGAACACAAGCCACGGGCUUCCUCUUCAAGAACUGGCAUCAAUUAUU ..((((((((.(((......((((((((((((..........))))).))).))))......((.....))......))).)))))))).......................... ( -24.10) >consensus CUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG_GAUCAGAAAGAACACAAGCCACGGGCUUCCUCUUCAAGAGCUGGCAUCAAUUAUU ..((((((((.(((......((((((((((((..........))))).))).))))......((.....))......))).)))))))).......................... (-21.64 = -22.04 + 0.40)


| Location | 13,760,106 – 13,760,224 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13760106 118 + 23771897 GCUUGUGUUCUUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAGAUGUUGAACGCCAGCCGUGGCUGUGCUUUUAUUGUUUAUU ((..((((((..(((((..-...((((.(((.((((((........)))))))))))))......(((...))).)))))....))))))((((....)))).)).............. ( -29.00) >DroSec_CAF1 6672 118 + 1 GCUUGUGUUCCUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAGAUGUUGAACGCCAGCCGUUGCUGUGCUUUUAUUGUUUAUU ((..((((((..(((((..-...((((.(((.((((((........)))))))))))))......(((...))).)))))....))))))((((....)))).)).............. ( -26.20) >DroSim_CAF1 2805 118 + 1 GCUUGUGUUCUUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAGAUGUUGAACGCCAGCCGUGGCUGUGCUUUUAUUGUUUAUU ((..((((((..(((((..-...((((.(((.((((((........)))))))))))))......(((...))).)))))....))))))((((....)))).)).............. ( -29.00) >DroEre_CAF1 5835 118 + 1 GCUUGUGUUCUUUCUGAUC-CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCCGGCUUCAGAUGUUGAACGCCAGCCGUGGCUGUGCUUUUAUUGUUUAUU ((..((((((..(((((..-...((((.(((.((((((........)))))))))))))......(((...))).)))))....))))))((((....)))).)).............. ( -29.00) >DroYak_CAF1 2909 119 + 1 GCUUGUGUUCUUUCUGAUCCCUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAGAUGUUGAACGCCAGCCGUGGCUGUGCUUUUAUUGUUUAUU ((..((((((..(((((..(((.....((((((((.(((..((.((......)).))))).)))))))).)))..)))))....))))))((((....)))).)).............. ( -29.80) >consensus GCUUGUGUUCUUUCUGAUC_CUUUUUAUGGUAUUAAGUUAUUAUGUGCUUAAACCUAAAUAUAAUGCCGCAGGCUUCAGAUGUUGAACGCCAGCCGUGGCUGUGCUUUUAUUGUUUAUU ((..((((((..(((((......((((.(((.((((((........)))))))))))))......(((...))).)))))....))))))((((....)))).)).............. (-28.44 = -28.44 + -0.00)



| Location | 13,760,106 – 13,760,224 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.81 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13760106 118 - 23771897 AAUAAACAAUAAAAGCACAGCCACGGCUGGCGUUCAACAUCUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAAGAACACAAGC ..............((.((((....))))))((((....(((((.(((...)))......((((((((((((..........))))).))).))))...-..)))))..))))...... ( -25.80) >DroSec_CAF1 6672 118 - 1 AAUAAACAAUAAAAGCACAGCAACGGCUGGCGUUCAACAUCUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAGGAACACAAGC ..............((.((((....))))))((((....(((((.(((...)))......((((((((((((..........))))).))).))))...-..)))))..))))...... ( -25.40) >DroSim_CAF1 2805 118 - 1 AAUAAACAAUAAAAGCACAGCCACGGCUGGCGUUCAACAUCUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAAGAACACAAGC ..............((.((((....))))))((((....(((((.(((...)))......((((((((((((..........))))).))).))))...-..)))))..))))...... ( -25.80) >DroEre_CAF1 5835 118 - 1 AAUAAACAAUAAAAGCACAGCCACGGCUGGCGUUCAACAUCUGAAGCCGGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG-GAUCAGAAAGAACACAAGC ..............((.((((....))))))((((....(((((.(((...)))......((((((((((((..........))))).))).))))...-..)))))..))))...... ( -25.80) >DroYak_CAF1 2909 119 - 1 AAUAAACAAUAAAAGCACAGCCACGGCUGGCGUUCAACAUCUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAGGGAUCAGAAAGAACACAAGC ..............((.((((....))))))((((....(((((.(((...)))......((((((((((((..........))))).))).))))......)))))..))))...... ( -25.80) >consensus AAUAAACAAUAAAAGCACAGCCACGGCUGGCGUUCAACAUCUGAAGCCUGCGGCAUUAUAUUUAGGUUUAAGCACAUAAUAACUUAAUACCAUAAAAAG_GAUCAGAAAGAACACAAGC ..............((.((((....))))))((((....(((((.(((...)))......((((((((((((..........))))).))).))))......)))))..))))...... (-25.72 = -25.72 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:21 2006