
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,759,088 – 13,759,367 |
| Length | 279 |
| Max. P | 0.997459 |

| Location | 13,759,088 – 13,759,207 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13759088 119 + 23771897 CCACUUCGCCGUU-GCCCCACAACUCGCUAAAUUUAUUAACCCACAUUAACCACUUCCAUCGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCU .......((.(((-(.....))))..)).................................(((((((((((((..((((((((.......))))).))).))))))))).....)))). ( -27.50) >DroSec_CAF1 5677 119 + 1 CCACUCCAACGUU-GCCACACAACUCGCUAAAUUUAUUAACCCACAUUAACCACUUCCAUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCU ......((((.((-((((.........................................))))))(((((((((..((((((((.......))))).))).)))))))))....)))).. ( -28.98) >DroSim_CAF1 1815 119 + 1 CCACUCGGACGUU-GCCACACAACUCGCUAAAUUUAUUAACCCACAUUAACCACUUCCAUGGCAAAAUGUUGAUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCU ......(((.(((-(.....))))..................(((((((.....(((((((((((....))).))))))))......)))))))..)))..((((((.......)))))) ( -25.20) >DroEre_CAF1 4905 119 + 1 CCACUCCGCGGUU-GCCACACAACUCCCUAAAUUUAUUAACCCACAUUAACCACUUUCCUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCU .......((((((-(.....))))).....(((((((.............(((......)))...(((((((((..((((((((.......))))).))).)))))))))))))))))). ( -29.10) >DroYak_CAF1 1933 120 + 1 CCACUCCGCGGUUCACCCCACAGCUCCCUAAAUUUAUUAACCCACAUUAACCACUUUCCUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCU .......((((......))...........(((((((.............(((......)))...(((((((((..((((((((.......))))).))).)))))))))))))))))). ( -27.30) >consensus CCACUCCGCCGUU_GCCACACAACUCGCUAAAUUUAUUAACCCACAUUAACCACUUCCAUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCU ....................(((((.........................(((......)))...(((((((((..((((((((.......))))).))).)))))))))...))))).. (-23.52 = -23.76 + 0.24)

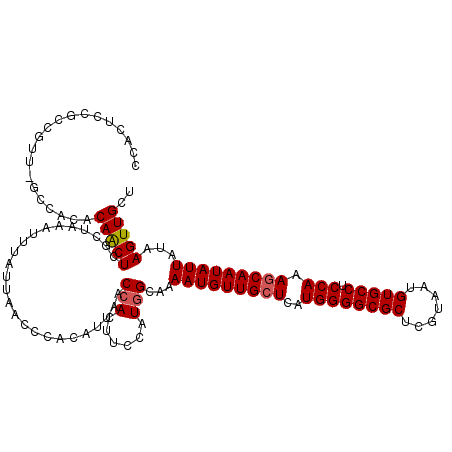

| Location | 13,759,127 – 13,759,247 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -29.48 |
| Energy contribution | -30.24 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13759127 120 + 23771897 CCCACAUUAACCACUUCCAUCGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCUGUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCG ................((((.(((((((((((((..((((((((.......))))).))).))))))))).....)))).)).))..............(((((((.....))))))).. ( -31.60) >DroSec_CAF1 5716 120 + 1 CCCACAUUAACCACUUCCAUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCUGUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCG ................((((((((((((((((((..((((((((.......))))).))).))))))))).....))))))).))..............(((((((.....))))))).. ( -34.50) >DroSim_CAF1 1854 120 + 1 CCCACAUUAACCACUUCCAUGGCAAAAUGUUGAUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCUGUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCG ...((((((.....(((((((((((....))).))))))))......))))))((((....((((((.......))))))..)))).............(((((((.....))))))).. ( -31.30) >DroEre_CAF1 4944 120 + 1 CCCACAUUAACCACUUUCCUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCUGUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAACUGCG ...........((.((((((((((((((((((((..((((((((.......))))).))).))))))))).....))))).....................((....)))))))).)).. ( -29.90) >DroYak_CAF1 1973 120 + 1 CCCACAUUAACCACUUUCCUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCUGUAGGCUUUUAAAACUUUUCACUUUCCAGAGGAAAGUGCG ...........(((((((((((((((((((((((..((((((((.......))))).))).))))))))).....)))))...((....................))..))))))))).. ( -35.95) >consensus CCCACAUUAACCACUUCCAUGGCAAAAUGUUGCUCAUGGGGCGCUCGUAAUGUGCCUCCAAAGCAAUAUUAUAAGUUGCUGUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCG ................((((((((((((((((((..((((((((.......))))).))).))))))))).....))))))).))..............(((((((.....))))))).. (-29.48 = -30.24 + 0.76)


| Location | 13,759,127 – 13,759,247 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -32.00 |
| Energy contribution | -32.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13759127 120 - 23771897 CGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUACAGCAACUUAUAAUAUUGCUUUGGAGGCACAUUACGAGCGCCCCAUGAGCAACAUUUUGCGAUGGAAGUGGUUAAUGUGGG ..(((((((.....)))))))..............((((((..(((........(((((((((.(((.(.......).)))))).))))))((((((......)))))))))..)))))) ( -33.70) >DroSec_CAF1 5716 120 - 1 CGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUACAGCAACUUAUAAUAUUGCUUUGGAGGCACAUUACGAGCGCCCCAUGAGCAACAUUUUGCCAUGGAAGUGGUUAAUGUGGG ..(((((((.....)))))))..............((((((..(((........(((((((((.(((.(.......).)))))).))))))((((((......)))))))))..)))))) ( -33.70) >DroSim_CAF1 1854 120 - 1 CGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUACAGCAACUUAUAAUAUUGCUUUGGAGGCACAUUACGAGCGCCCCAUGAUCAACAUUUUGCCAUGGAAGUGGUUAAUGUGGG ..(((((((.....))))))).............((((..(((((.........)))))....))))((((((...((...(((((..(((....))).)))))..))...))))))... ( -27.80) >DroEre_CAF1 4944 120 - 1 CGCAGUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUACAGCAACUUAUAAUAUUGCUUUGGAGGCACAUUACGAGCGCCCCAUGAGCAACAUUUUGCCAGGAAAGUGGUUAAUGUGGG .........((.((((((.(....).)))))).))((((((..(((........(((((((((.(((.(.......).)))))).))))))((((((......)))))))))..)))))) ( -30.40) >DroYak_CAF1 1973 120 - 1 CGCACUUUCCUCUGGAAAGUGAAAAGUUUUAAAAGCCUACAGCAACUUAUAAUAUUGCUUUGGAGGCACAUUACGAGCGCCCCAUGAGCAACAUUUUGCCAGGAAAGUGGUUAAUGUGGG ..((((((((...))))))))..............((((((..(((........(((((((((.(((.(.......).)))))).))))))((((((......)))))))))..)))))) ( -36.10) >consensus CGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUACAGCAACUUAUAAUAUUGCUUUGGAGGCACAUUACGAGCGCCCCAUGAGCAACAUUUUGCCAUGGAAGUGGUUAAUGUGGG ..(((((((.....)))))))..............((((((..(((........(((((((((.(((.(.......).)))))).))))))((((((......)))))))))..)))))) (-32.00 = -32.16 + 0.16)


| Location | 13,759,207 – 13,759,327 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.38 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13759207 120 + 23771897 GUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCGGUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUC ...((((......(((((((.((....)).)))))))((((((.((((.......)))((((((......(((((((.....)))..)))).....))))))......).)))))))))) ( -31.20) >DroSec_CAF1 5796 120 + 1 GUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCGGUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUC ...((((......(((((((.((....)).)))))))((((((.((((.......)))((((((......(((((((.....)))..)))).....))))))......).)))))))))) ( -31.20) >DroSim_CAF1 1934 120 + 1 GUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCGGUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUC ...((((......(((((((.((....)).)))))))((((((.((((.......)))((((((......(((((((.....)))..)))).....))))))......).)))))))))) ( -31.20) >DroEre_CAF1 5024 120 + 1 GUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAACUGCGGUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUC .................(((.(((....))))))(((((((((.((((.......)))((((((......(((((((.....)))..)))).....))))))......).))))))))). ( -30.10) >DroYak_CAF1 2053 120 + 1 GUAGGCUUUUAAAACUUUUCACUUUCCAGAGGAAAGUGCGGUUUCGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCAGUGACCGCAGUC ...((((.............(((((((...)))))))((((((.((((.......)))((((((......(((((((.....)))..)))).....))))))......).)))))))))) ( -32.40) >consensus GUAGGCUUUUAAAACUUUUCACUUUCAAGAGGAAAGUGCGGUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUC ...((((......(((((((.((....)).)))))))((((((.((((.......)))((((((......(((((((.....)))..)))).....))))))......).)))))))))) (-30.50 = -30.38 + -0.12)



| Location | 13,759,207 – 13,759,327 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -30.66 |
| Consensus MFE | -28.60 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13759207 120 - 23771897 GACUGCGGUCAGUGGGUUGGCAAAAAUGAUUCAUUAGCAUUCAGCUUGAAAGUUGAUUUGCCGCAAAAGAGAUACCAAACCGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUAC (((....))).(((((((((((((((((...)))).....((((((....))))))))))))..............((((..(((((((.....)))))))....))))....))))))) ( -30.00) >DroSec_CAF1 5796 120 - 1 GACUGCGGUCAGUGGGUUGGCAAAAAUGAUUCAUUAGCAUUCAGCUUGAAAGUUGAUUUGCCGCAAAAGAGAUACCAAACCGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUAC (((....))).(((((((((((((((((...)))).....((((((....))))))))))))..............((((..(((((((.....)))))))....))))....))))))) ( -30.00) >DroSim_CAF1 1934 120 - 1 GACUGCGGUCAGUGGGUUGGCAAAAAUGAUUCAUUAGCAUUCAGCUUGAAAGUUGAUUUGCCGCAAAAGAGAUACCAAACCGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUAC (((....))).(((((((((((((((((...)))).....((((((....))))))))))))..............((((..(((((((.....)))))))....))))....))))))) ( -30.00) >DroEre_CAF1 5024 120 - 1 GACUGCGGUCAGUGGGUUGGCAAAAAUGAUUCAUUAGCAUUCAGCUUGAAAGUUGAUUUGCCGCAAAAGAGAUACCAAACCGCAGUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUAC (((((((((...(((...((((((((((...)))).....((((((....)))))))))))).(......)...))).)))))))))..((.((((((.(....).)))))).))..... ( -31.30) >DroYak_CAF1 2053 120 - 1 GACUGCGGUCACUGGGUUGGCAAAAAUGAUUCAUUAGCAUUCAGCUUGAAAGUUGAUUUGCCGCAAAAGAGAUACGAAACCGCACUUUCCUCUGGAAAGUGAAAAGUUUUAAAAGCCUAC (((....)))..((((((((((((((((...)))).....((((((....)))))))))))).............(((((..((((((((...))))))))....)))))...)))))). ( -32.00) >consensus GACUGCGGUCAGUGGGUUGGCAAAAAUGAUUCAUUAGCAUUCAGCUUGAAAGUUGAUUUGCCGCAAAAGAGAUACCAAACCGCACUUUCCUCUUGAAAGUGAAAAGUUUUAAAAGCCUAC (((....))).(((((((((((((((((...)))).....((((((....))))))))))))..............((((..(((((((.....)))))))....))))....))))))) (-28.60 = -29.00 + 0.40)

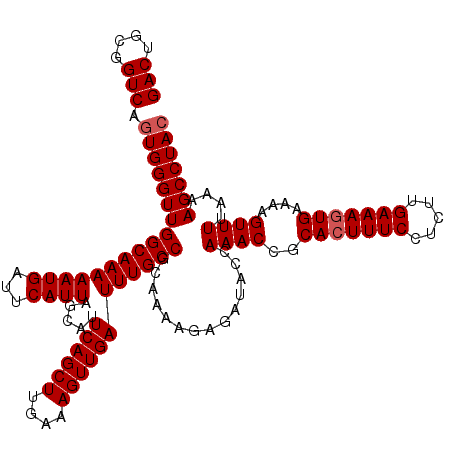
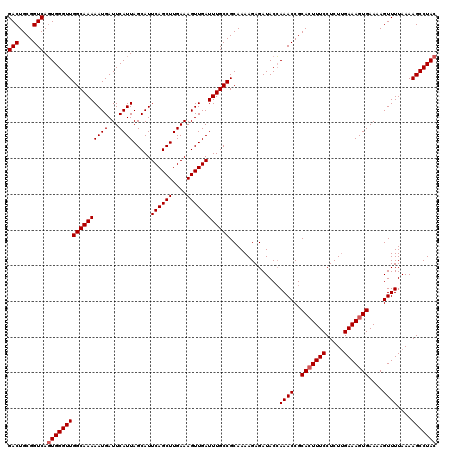
| Location | 13,759,247 – 13,759,367 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13759247 120 + 23771897 GUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUCGGUAAAGGUAGCGUUAGUUGGCCCAUACCAUUUAUUAGUG ...((((((.........((((((......(((((((.....)))..)))).....))))))..((.((((((.((..((......))..)))))))).))...)))))).......... ( -30.20) >DroSec_CAF1 5836 120 + 1 GUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUCGGUAAAGGUAGCGUUAGUUGGCCCAUACCAUUUAUUAGUG ...((((((.........((((((......(((((((.....)))..)))).....))))))..((.((((((.((..((......))..)))))))).))...)))))).......... ( -30.20) >DroSim_CAF1 1974 120 + 1 GUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUCGGUAAAGGUAGCGUAAGUUGGCCCAUACCAUUUAUUAGUG ...((((((((..(((((((((((......(((((((.....)))..)))).....)))))).(((.((((((....))))))...)))...)))))..))...)))))).......... ( -31.00) >DroEre_CAF1 5064 120 + 1 GUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUCGGUAAAGGUAGCGUUAGUUGGCCCAUACCAUUUAUUGGUG ....(((....((..(((((((((......(((((((.....)))..)))).....)))))).(((.((((((....))))))...))).)))..))...)))..(((((.....))))) ( -33.60) >DroYak_CAF1 2093 116 + 1 GUUUCGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCAGUGACCGCAGUCGGUAAAGGUAGCGUUACUUGG----UAUCAUUUAUUAGUG .....(((.......)))((((((......(((((((.....)))..)))).....)))))).(((.((((((.((..((......))..)))))))).))----).............. ( -27.10) >consensus GUUUGGUAUCUCUUUUGCGGCAAAUCAACUUUCAAGCUGAAUGCUAAUGAAUCAUUUUUGCCAACCCACUGACCGCAGUCGGUAAAGGUAGCGUUAGUUGGCCCAUACCAUUUAUUAGUG ...((((((.........((((((......(((((((.....)))..)))).....))))))..((.((((((.((..((......))..)))))))).))...)))))).......... (-28.00 = -28.12 + 0.12)

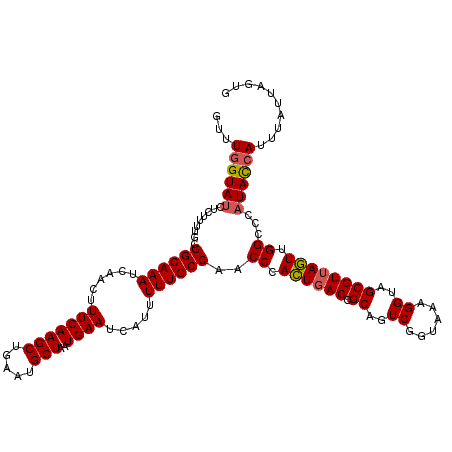

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:17 2006