
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,756,404 – 13,756,566 |
| Length | 162 |
| Max. P | 0.988253 |

| Location | 13,756,404 – 13,756,494 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
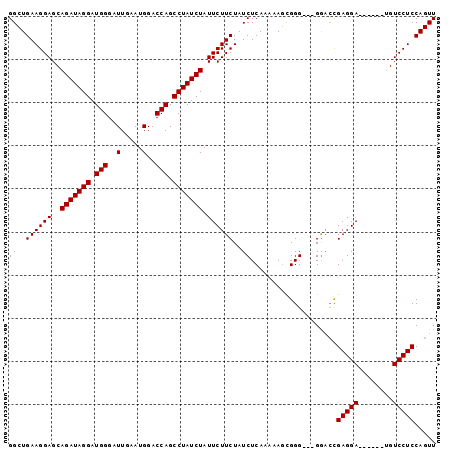
>3L_DroMel_CAF1 13756404 90 + 23771897 GGCUGAAGGAGCAGAUAGGAUGGGAUUGAAUGGACCAGCCUAUCUAUUCUUCUAUCUCAAAAAGCGGG---GAGCCGAGGA------UGUCCUCCAGUU ((((((((((..(((((((.(((..(.....)..))).))))))).))))))..((((.......)))---)))))((((.------...))))..... ( -33.40) >DroSim_CAF1 175391 90 + 1 GGCUGAAGGAGCAGAUAGGAUGGGAUUGAAUGGACCAGCCUAUCUAUUCUUCUAUCUCAAAAAUCGGG---GGACCGAGGA------UGUCCUCCAGUU ((..((((((..(((((((.(((..(.....)..))).))))))).))))))..)).........(((---((((......------.))))))).... ( -32.50) >DroYak_CAF1 174366 99 + 1 GGCUGAAGGAGCAGAUAGGAUGGGAUUGAAUGGACCAGCCUAUCUAUUCUUCUAUCUCAAAAAGUGGGCGAGGAGUGAGGAGGAGUGAGUCCUCCAGUU .(((.....)))(((((((.(((..(.....)..))).)))))))(((((((...((((.....)))).)))))))..((((((.....)))))).... ( -33.30) >consensus GGCUGAAGGAGCAGAUAGGAUGGGAUUGAAUGGACCAGCCUAUCUAUUCUUCUAUCUCAAAAAGCGGG___GGACCGAGGA______UGUCCUCCAGUU ((..((((((..(((((((.(((..(.....)..))).))))))).))))))..))....................(((((........)))))..... (-26.47 = -26.47 + -0.00)


| Location | 13,756,404 – 13,756,494 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -24.83 |
| Energy contribution | -24.94 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
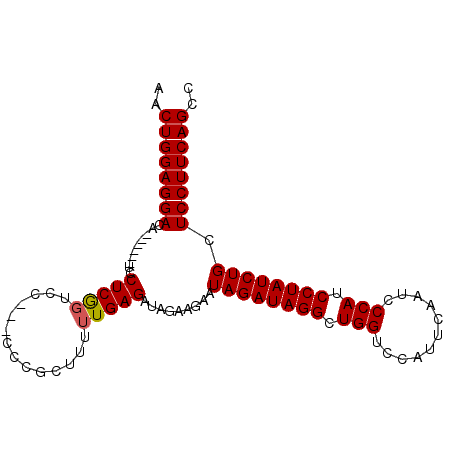
>3L_DroMel_CAF1 13756404 90 - 23771897 AACUGGAGGACA------UCCUCGGCUC---CCCGCUUUUUGAGAUAGAAGAAUAGAUAGGCUGGUCCAUUCAAUCCCAUCCUAUCUGCUCCUUCAGCC ..((((((((..------..((((((..---...)))....))).........((((((((.(((...........))).)))))))).)))))))).. ( -26.60) >DroSim_CAF1 175391 90 - 1 AACUGGAGGACA------UCCUCGGUCC---CCCGAUUUUUGAGAUAGAAGAAUAGAUAGGCUGGUCCAUUCAAUCCCAUCCUAUCUGCUCCUUCAGCC ..((((((((((------...((((...---.))))....))...........((((((((.(((...........))).)))))))).)))))))).. ( -27.30) >DroYak_CAF1 174366 99 - 1 AACUGGAGGACUCACUCCUCCUCACUCCUCGCCCACUUUUUGAGAUAGAAGAAUAGAUAGGCUGGUCCAUUCAAUCCCAUCCUAUCUGCUCCUUCAGCC ....((((((.....))))))......((((.........))))...((((..((((((((.(((...........))).))))))))...)))).... ( -26.70) >consensus AACUGGAGGACA______UCCUCGGUCC___CCCGCUUUUUGAGAUAGAAGAAUAGAUAGGCUGGUCCAUUCAAUCCCAUCCUAUCUGCUCCUUCAGCC ..((((((((..........(((((..............))))).........((((((((.(((...........))).)))))))).)))))))).. (-24.83 = -24.94 + 0.11)

| Location | 13,756,465 – 13,756,566 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -21.69 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
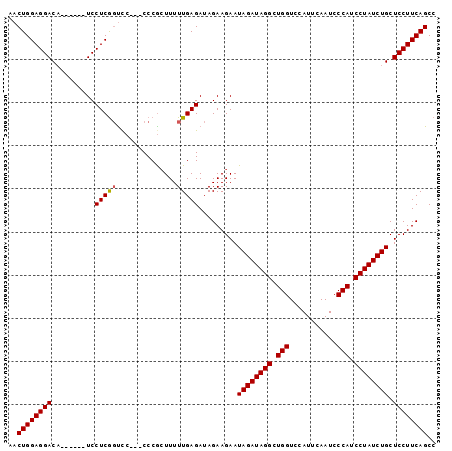
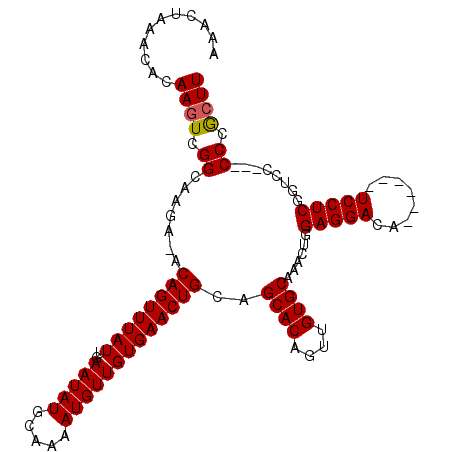
>3L_DroMel_CAF1 13756465 101 - 23771897 AAACUAAACACAAGUCGGCAAGA-ACAGUUUAUUGAAUAUGCAAAAUGUUGUGAACUGCAGCACAGUUGUGCAAACUGGAGGACA------UCCUCGGCUC---CCCGCUU ...........(((.(((.....-.((((((((..(((((.....))))))))))))).(((.(((((.....)))))((((...------.)))).))).---.)))))) ( -25.90) >DroSim_CAF1 175452 101 - 1 AAACUAAACACAAGUCGGCAAGA-ACAGUUUAUUGAAUAUGCAAAAUGUUGUGAACUGCAGCACAGUUGUGCAAACUGGAGGACA------UCCUCGGUCC---CCCGAUU ............((((((.....-.((((((((..(((((.....)))))))))))))..((((....))))..((((.(((...------.)))))))..---.)))))) ( -25.80) >DroYak_CAF1 174427 111 - 1 AAACUAAACACAAGUCGGCAAGAAACAGUUUAUUGAAUAUGCAAAAUGUUGUGAACUGCAGCACAGUUGUGCAAACUGGAGGACUCACUCCUCCUCACUCCUCGCCCACUU ...........((((.(((.((...((((((((..(((((.....)))))))))))))..((((....)))).....((((((.....))))))......)).))).)))) ( -31.40) >consensus AAACUAAACACAAGUCGGCAAGA_ACAGUUUAUUGAAUAUGCAAAAUGUUGUGAACUGCAGCACAGUUGUGCAAACUGGAGGACA______UCCUCGGUCC___CCCGCUU ...........((((.((.......((((((((..(((((.....)))))))))))))..((((....))))......(((((........)))))........)).)))) (-21.69 = -21.80 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:10 2006