| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,402,833 – 1,402,939 |
| Length | 106 |
| Max. P | 0.630428 |

| Location | 1,402,833 – 1,402,939 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -17.52 |
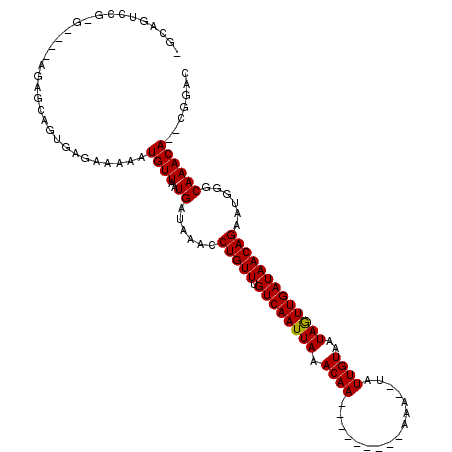
| Consensus MFE | -11.05 |
| Energy contribution | -10.91 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1402833 106 - 23771897 UGGAGUCCGUGGCCAAGAGCAGUGAGAAAAAUGUUAAUGAUAAACCUGUUUGUCAAUUAAACAA--------AAAA--UAUUGUAAUAGUUGAUAACAGAAUGGGCAAACA--CGAAG ....((((((...((..((((..........))))..))......(((((.((((((((.((((--------....--..))))..))))))))))))).)))))).....--..... ( -21.70) >DroPse_CAF1 85466 105 - 1 AGCAGAUCGAG----AGAACAGUGAGAAAAAUGUUAAUGAUAAACCUGUUUGUCAAUUAAACAA---------AAAUAUAUUGUAAUAGUUGAUAACAGACUAAGCAAACAGACGGAC .((..((((..----..((((..........))))..))))....(((((.((((((((.((((---------.......))))..))))))))))))).....))............ ( -16.30) >DroEre_CAF1 59368 92 - 1 UGGAGUC-------------AGUGAGAAAAAUGUUAAUGAUAAACCUGUUUGUCAAUUAAACAA---------AAA--UAUUGUAAUAAUUGAUAACAGAAUGGGCAAACA--CGAAC ....(((-------------(.(((.(....).))).))))....(((((.((((((((.((((---------...--..))))..)))))))))))))............--..... ( -14.10) >DroWil_CAF1 37888 106 - 1 --------UCCAUCUGAUGCAGUGAGAAAAAUGUUAAUGAUAAACCUGUUUGUCAAUUAAACAAAAUACAAAAAAA--UAUUGUAAUAGUUGAUAACAGAAUGGGCAAACA--GUGCC --------.(((((((..((((.(......((....))......)))))((((((((((.((((.((........)--).))))..))))))))))))).))))(((....--.))). ( -17.10) >DroAna_CAF1 61967 96 - 1 --AAGUCC-------AGGGCAGUGAGAAAAAUGUUAAUGAUAAACCUGUUUGUCAAUUAAACAA---------AAA--UAUUGUAAUAGUUGAUAACAGAAUGGGCAAACA--CGGAG --..((((-------(.((((..........))))..........(((((.((((((((.((((---------...--..))))..)))))))))))))..))))).....--..... ( -19.60) >DroPer_CAF1 86837 105 - 1 AGCAGAUCGAG----AGAACAGUGAGAAAAAUGUUAAUGAUAAACCUGUUUGUCAAUUAAACAA---------AAAUAUAUUGUAAUAGUUGAUAACAGACUAAGCAAACAGACGGAC .((..((((..----..((((..........))))..))))....(((((.((((((((.((((---------.......))))..))))))))))))).....))............ ( -16.30) >consensus _GCAGUCCG_G____AGAGCAGUGAGAAAAAUGUUAAUGAUAAACCUGUUUGUCAAUUAAACAA_________AAA__UAUUGUAAUAGUUGAUAACAGAAUGGGCAAACA__CGGAC ...............................((((..((......(((((.((((((((.((((................))))..)))))))))))))......))))))....... (-11.05 = -10.91 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:37 2006