
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,753,900 – 13,754,108 |
| Length | 208 |
| Max. P | 0.974807 |

| Location | 13,753,900 – 13,754,003 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -32.95 |
| Energy contribution | -34.20 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13753900 103 + 23771897 CUUAAAUUCCUCUUGGCCAGGAGAAGGACACGGGGCAAUCUCACUUUCCCGCUUACACUCUCUGCUGGUUAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUC ..............(((((((.(((((....(((.....))).)))))))((((.((((((((((..(...)..))))))))))...)))).)))))...... ( -38.60) >DroSim_CAF1 172883 103 + 1 CUUAAAUUCCUCUUGGCCAGGAGAAGGACACGGGGCAAUCUCACUUGCCCGCUUACACUCUCUGCUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUC .........(((((....)))))..((.(((.((((((......))))))((((.((((((((((..(...)..))))))))))...))))))).))...... ( -42.30) >DroEre_CAF1 167461 103 + 1 CUUAAAUUCCUCUUGGCCAGCAAAAGGACACGGGUCCAUCUGACUUUCCCGCUUACACUCUCUGCUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUC ..............(((((......((((....))))............(((((.((((((((((..(...)..))))))))))...))))))))))...... ( -39.40) >DroYak_CAF1 171869 103 + 1 CUUAAAUUCCUCUUGGCCAGCAGAAGGACACGGGGCCAUCUCACUUUCCCGCUUACACUCUCGACUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUC .......((((..((.....))..)))).(((((((((...((((((...((...(((((((.....(((...))).)))))))))))))))))))).)))). ( -33.00) >consensus CUUAAAUUCCUCUUGGCCAGCAGAAGGACACGGGGCAAUCUCACUUUCCCGCUUACACUCUCUGCUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUC ..............(((((.((((.((........)).)))).......(((((.((((((((((..(...)..))))))))))...))))))))))...... (-32.95 = -34.20 + 1.25)

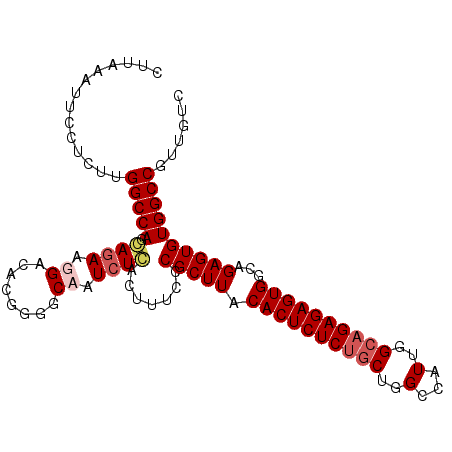

| Location | 13,753,900 – 13,754,003 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -31.88 |
| Energy contribution | -32.75 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13753900 103 - 23771897 GACAACGGCCACACUCUGCCACUCUCUGCCAAUAACCAGCAGAGAGUGUAAGCGGGAAAGUGAGAUUGCCCCGUGUCCUUCUCCUGGCCAAGAGGAAUUUAAG ......(((((......((((((((((((.........))))))))))...))((..(((.((.((......)).)))))..))))))).............. ( -34.30) >DroSim_CAF1 172883 103 - 1 GACAACGGCCACACUCUGCCACUCUCUGCCAAUGGCCAGCAGAGAGUGUAAGCGGGCAAGUGAGAUUGCCCCGUGUCCUUCUCCUGGCCAAGAGGAAUUUAAG ......((((((((...((((((((((((.........))))))))))...))((((((......)))))).)))).........)))).............. ( -37.30) >DroEre_CAF1 167461 103 - 1 GACAACGGCCACACUCUGCCACUCUCUGCCAAUGGCCAGCAGAGAGUGUAAGCGGGAAAGUCAGAUGGACCCGUGUCCUUUUGCUGGCCAAGAGGAAUUUAAG ......((((....(((((((((((((((.........))))))))))...)))))..((.((((.((((....)))).)))))))))).............. ( -38.40) >DroYak_CAF1 171869 103 - 1 GACAACGGCCACACUCUGCCACUCUCUGCCAAUGGCCAGUCGAGAGUGUAAGCGGGAAAGUGAGAUGGCCCCGUGUCCUUCUGCUGGCCAAGAGGAAUUUAAG ((((..((((((((((((((((((((.(((...))).....)))))))...))))...))))...)))))...))))(((((........)))))........ ( -35.10) >consensus GACAACGGCCACACUCUGCCACUCUCUGCCAAUGGCCAGCAGAGAGUGUAAGCGGGAAAGUGAGAUGGCCCCGUGUCCUUCUCCUGGCCAAGAGGAAUUUAAG ((((..(((((((((((((((((((((((.........))))))))))...))))...))))...)))))...))))(((((........)))))........ (-31.88 = -32.75 + 0.88)



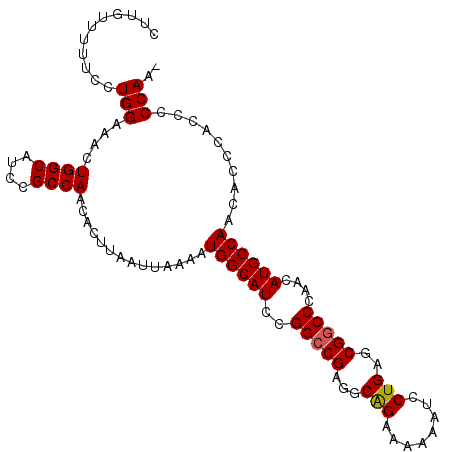
| Location | 13,753,926 – 13,754,043 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -43.13 |
| Consensus MFE | -40.44 |
| Energy contribution | -40.95 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13753926 117 + 23771897 GACACGGGGCAAUCUCACUUUCCCGCUUACACUCUCUGCUGGUUAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUCCUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUU ....(((((...........)))))....((((((((((..(...)..))))))))))...(((((((((.(.((((...(((((.....))))).)))).).))).)))))).... ( -43.40) >DroSim_CAF1 172909 117 + 1 GACACGGGGCAAUCUCACUUGCCCGCUUACACUCUCUGCUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUCCUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUU ......((((((......))))))((...((((((((((..(...)..)))))))))))).(((((((((.(.((((...(((((.....))))).)))).).))).)))))).... ( -48.40) >DroEre_CAF1 167487 117 + 1 GACACGGGUCCAUCUGACUUUCCCGCUUACACUCUCUGCUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUCCUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUU ....((((.............))))....((((((((((..(...)..))))))))))...(((((((((.(.((((...(((((.....))))).)))).).))).)))))).... ( -43.02) >DroYak_CAF1 171895 117 + 1 GACACGGGGCCAUCUCACUUUCCCGCUUACACUCUCGACUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUCCUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUU ....(((((...........)))))....(((((((.....(((...))).)))))))...(((((((((.(.((((...(((((.....))))).)))).).))).)))))).... ( -37.70) >consensus GACACGGGGCAAUCUCACUUUCCCGCUUACACUCUCUGCUGGCCAUUGGCAGAGAGUGGCAGAGUGUGGCCGUUGUCCUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUU ....((((.............))))....((((((((((..(...)..))))))))))...(((((((((.(.((((...(((((.....))))).)))).).))).)))))).... (-40.44 = -40.95 + 0.50)

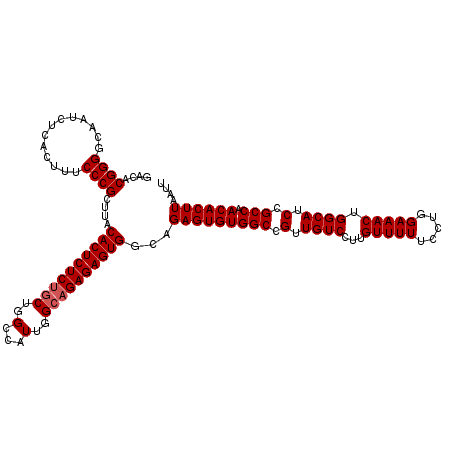

| Location | 13,754,003 – 13,754,108 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 96.21 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13754003 105 + 23771897 CUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUUAAAAUGGCAUCCGGGCGAGGCAGAAAAAAUCCUGAGCGGCCCAACAUGCCAACACCCAUCCCCAA- ...........(((....((((....))))..............((((((..((((....(((........)))....))))...))))))..........))).- ( -26.20) >DroSim_CAF1 172986 105 + 1 CUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUUAAAAUGGCAUCCGGCCGAGGCAGAAAAAAUCCUGAGCGGCCCAACAUGCCAACACCCACCCCCAA- ...........(((....((((....))))..............((((((..(((((...(((........)))..)))))....))))))..........))).- ( -29.00) >DroEre_CAF1 167564 106 + 1 CUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUUAAAAUGGCAUCCGGCCGAGGCGGAAAAAAUCCUGAGCGGCCCAACAUGCCAACAACCACCUCCAAC ...........((((...((((....))))..............((((((..(((((...(((........)))..)))))....)))))).........)))).. ( -29.70) >consensus CUUGUUUUUCCUGGAAACUGGCAUCCGCCAACACUUAAUUAAAAUGGCAUCCGGCCGAGGCAGAAAAAAUCCUGAGCGGCCCAACAUGCCAACACCCACCCCCAA_ ...........(((....((((....))))..............((((((..(((((...(((........)))..)))))....))))))..........))).. (-26.35 = -26.47 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:03 2006