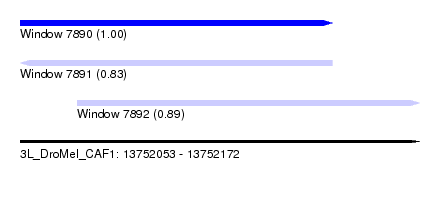
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,752,053 – 13,752,172 |
| Length | 119 |
| Max. P | 0.998538 |

| Location | 13,752,053 – 13,752,146 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.76 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13752053 93 + 23771897 GUUAGCAAAUUGCCUGGGUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGCA---- ((((.((((((((((.(((((.((((...((((((.....)).))))))))..))))).))))((((((......)))))))))))).)))).---- ( -37.30) >DroSec_CAF1 158791 93 + 1 GUUAGCAAAUUGCCUGGGUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGUG---- .(((.((((((((((.(((((.((((...((((((.....)).))))))))..))))).))))((((((......)))))))))))).)))..---- ( -33.40) >DroSim_CAF1 171028 93 + 1 GUUAGCAAAUUGCCUGGGUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGUG---- .(((.((((((((((.(((((.((((...((((((.....)).))))))))..))))).))))((((((......)))))))))))).)))..---- ( -33.40) >DroEre_CAF1 165646 93 + 1 GUUAGCAAAUUGCCUGGGUCUUGCAGCAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGAUGGCUGAUUUGGUAGUG---- .(((.((((((((((.(((.(((.((((..(((((.....)).)))..)))))))))).))))(((((........))))))))))).)))..---- ( -35.70) >DroYak_CAF1 170000 97 + 1 GUUAUCAAAUUGCCUGCGUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCCCAAACUGAGGCGGCCGGAACUGGAUGGCUGAUUUGAUAGCGAUUG (((((((((((((((((((..(......)..)))))..(.((..(((((((((.....).))))))))..)).)...))).)))))))))))..... ( -39.80) >consensus GUUAGCAAAUUGCCUGGGUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGUG____ ((((.((((((((((.(((((.((((...((((((.....)).))))))))..))))).))))(((((........))))))))))).))))..... (-31.60 = -31.76 + 0.16)


| Location | 13,752,053 – 13,752,146 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -22.02 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13752053 93 - 23771897 ----UGCUACCAAAUCAGCCAACCAGUUCCGGCCGCCUCAGUUUGAGCAGCCGCCACAGUUUGCGCGAUUUCUGCAAAACCCAGGCAAUUUGCUAAC ----(((....(((((.((.(((..((..((((.((.((.....)))).))))..)).))).))..)))))..))).......(((.....)))... ( -22.30) >DroSec_CAF1 158791 93 - 1 ----CACUACCAAAUCAGCCAACCAGUUCCGGCCGCCUCAGUUUGAGCAGCCGCCACAGUUUGCGCGAUUUCUGCAAAACCCAGGCAAUUUGCUAAC ----......(((((..(((.....((..((((.((.((.....)))).))))..))..((((((.(....))))))).....))).)))))..... ( -22.00) >DroSim_CAF1 171028 93 - 1 ----CACUACCAAAUCAGCCAACCAGUUCCGGCCGCCUCAGUUUGAGCAGCCGCCACAGUUUGCGCGAUUUCUGCAAAACCCAGGCAAUUUGCUAAC ----......(((((..(((.....((..((((.((.((.....)))).))))..))..((((((.(....))))))).....))).)))))..... ( -22.00) >DroEre_CAF1 165646 93 - 1 ----CACUACCAAAUCAGCCAUCCAGUUCCGGCCGCCUCAGUUUGAGCAGCCGCCACAGUUUGCGCGAUUGCUGCAAGACCCAGGCAAUUUGCUAAC ----......(((((..(((..........))).((((..((((..((((((((((.....)).)))...))))).))))..)))).)))))..... ( -24.70) >DroYak_CAF1 170000 97 - 1 CAAUCGCUAUCAAAUCAGCCAUCCAGUUCCGGCCGCCUCAGUUUGGGCAGCCGCCACAGUUUGCGCGAUUUCUGCAAAACGCAGGCAAUUUGAUAAC .......((((((((..(((.....((..((((.((((......)))).))))..)).((((..(((.....))).))))...))).)))))))).. ( -29.80) >consensus ____CACUACCAAAUCAGCCAACCAGUUCCGGCCGCCUCAGUUUGAGCAGCCGCCACAGUUUGCGCGAUUUCUGCAAAACCCAGGCAAUUUGCUAAC ..........(((((..(((.....((..((((.((.((.....)))).))))..))..((((((.(....))))))).....))).)))))..... (-22.02 = -21.70 + -0.32)


| Location | 13,752,070 – 13,752,172 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -38.89 |
| Energy contribution | -38.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13752070 102 + 23771897 GUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGCAGUUGCUGUUGGUGGCGUUGUAGCUGC .....((((..((.((((...(.(..(((((((((...((..((.(((((((......))))))).))..)))))))))))..).)...)))).))..)))) ( -42.60) >DroSec_CAF1 158808 102 + 1 GUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGUGGUUGCUGUUGGUGGCGUUGCAGCUGC .....((((.....((((.(((.(..((.(((((.((((.(.((.((((((....)))))).)).)))))))))).))..).)))....)))).....)))) ( -40.10) >DroSim_CAF1 171045 102 + 1 GUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGUGGUUGCUGUUGGUGGCGUUGCAGCUGC .....((((.....((((.(((.(..((.(((((.((((.(.((.((((((....)))))).)).)))))))))).))..).)))....)))).....)))) ( -40.10) >DroEre_CAF1 165663 102 + 1 GUCUUGCAGCAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGAUGGCUGAUUUGGUAGUGGUUGCUGUUGGUGGCGUUGCAGCUGC .....(((((....((((...(.(..(((((..((...((..((.((((((........)))))).))..))))..)))))..).)...))))....))))) ( -40.70) >consensus GUUUUGCAGAAAUCGCGCAAACUGUGGCGGCUGCUCAAACUGAGGCGGCCGGAACUGGUUGGCUGAUUUGGUAGUGGUUGCUGUUGGUGGCGUUGCAGCUGC .....((((.....((((.(((.(..((.(((((.((((.(.((.((((((....)))))).)).)))))))))).))..).)))....)))).....)))) (-38.89 = -38.95 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:59 2006