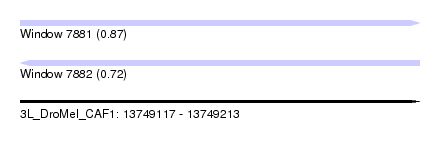
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,749,117 – 13,749,213 |
| Length | 96 |
| Max. P | 0.874658 |

| Location | 13,749,117 – 13,749,213 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -14.69 |
| Energy contribution | -16.72 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
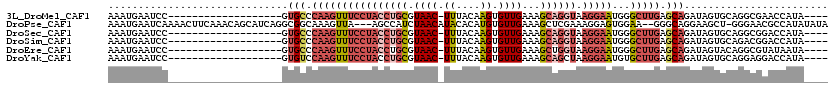
>3L_DroMel_CAF1 13749117 96 + 23771897 AAAUGAAUCC-------------------GUGCCCAAGUUUCCUACCUGCGUAAC-UUUACAAGUGUUGAAAGCAGGUAAGGAAUGGGCUUGAGCAGAUAGUGCAGGCGAACCAUA---- .........(-------------------(((((((...(((((((((((.((((-.........))))...)))))).))))))))))....(((.....)))..))).......---- ( -31.20) >DroPse_CAF1 158115 114 + 1 AAAUGAAUCAAAACUUCAAACAGCAUCAGGCGGCAAAGUUA---AGCCAUCUAACAUACACAUGUGUUGAAAGCUCGAAAGGAGUGGAA--GGGCAGGAAGCU-GGGAACGCCAUAUAUA ..............(((...((((.((..(((((.......---.))).(((((((((....))))))....((((.....))))))).--..))..)).)))-).)))........... ( -24.50) >DroSec_CAF1 155840 96 + 1 AAAUGAAUCC-------------------GUGCCCAAGUUUCCUACCUGCGUAAC-UUUACAAGUGUUGAAAGCAGGUAAGGAAUGGGCUUGAGCAGAUAGUGCAGGCGGACCAUA---- ..(((..(((-------------------(((((((...(((((((((((.((((-.........))))...)))))).))))))))))....(((.....)))..))))).))).---- ( -36.10) >DroSim_CAF1 167638 96 + 1 AAAUGAAUCC-------------------GUGCCCAAGUUUCCUACCUGCGUAAC-UUUACAAGUGUUGAAAGCAGGUAAGGAAUGGGCUUGAGCAGAUAGUGCAGACGGACCAUA---- ..(((..(((-------------------(((((((...(((((((((((.((((-.........))))...)))))).))))))))))....(((.....)))..))))).))).---- ( -35.80) >DroEre_CAF1 162718 96 + 1 AAAUGAAUCC-------------------GUGCCCAAGUUUCCUACCUGCGUAAC-UUUACAAGUGUUGAAAGCUGGUAAGGAAUGGGCUUGAGCAGAUAGUACAGGCGUAUAAUA---- ..........-------------------(((((((...((((((((.((.((((-.........))))...)).))).))))))))(((((.((.....)).)))))))))....---- ( -25.10) >DroYak_CAF1 167023 96 + 1 AAAUGAAUCC-------------------GUGUCCAAGUUUCCUACCUGCGUAAC-UUUACAAGUGUUGAAAGCAGCUAAGGAAUGUGCUUGAGCAGAUAGUGCAGGAGGACCAUA---- ..........-------------------(.((((....(((((..((((.((((-.........))))...))))...)))))((..((.........))..))...)))))...---- ( -22.90) >consensus AAAUGAAUCC___________________GUGCCCAAGUUUCCUACCUGCGUAAC_UUUACAAGUGUUGAAAGCAGGUAAGGAAUGGGCUUGAGCAGAUAGUGCAGGCGGACCAUA____ ..............................(((.((((((((((((((((.((((.((....)).))))...)))))).)))))...))))).)))........................ (-14.69 = -16.72 + 2.03)

| Location | 13,749,117 – 13,749,213 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -12.13 |
| Energy contribution | -14.72 |
| Covariance contribution | 2.58 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
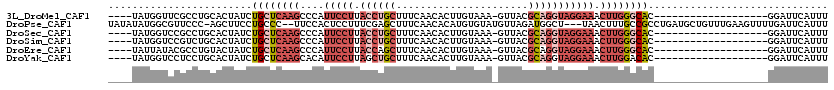
>3L_DroMel_CAF1 13749117 96 - 23771897 ----UAUGGUUCGCCUGCACUAUCUGCUCAAGCCCAUUCCUUACCUGCUUUCAACACUUGUAAA-GUUACGCAGGUAGGAAACUUGGGCAC-------------------GGAUUCAUUU ----.((((((((...(((.....)))....((((((((((.((((((....(((.........-)))..)))))))))))...))))).)-------------------))).)))).. ( -28.50) >DroPse_CAF1 158115 114 - 1 UAUAUAUGGCGUUCCC-AGCUUCCUGCCC--UUCCACUCCUUUCGAGCUUUCAACACAUGUGUAUGUUAGAUGGCU---UAACUUUGCCGCCUGAUGCUGUUUGAAGUUUUGAUUCAUUU ((((((((..(((...-(((((.......--.............)))))...))).)))))))).(((((.((((.---.......)))).)))))......((((.......))))... ( -20.75) >DroSec_CAF1 155840 96 - 1 ----UAUGGUCCGCCUGCACUAUCUGCUCAAGCCCAUUCCUUACCUGCUUUCAACACUUGUAAA-GUUACGCAGGUAGGAAACUUGGGCAC-------------------GGAUUCAUUU ----.((((((((...(((.....)))....((((((((((.((((((....(((.........-)))..)))))))))))...))))).)-------------------))).)))).. ( -31.40) >DroSim_CAF1 167638 96 - 1 ----UAUGGUCCGUCUGCACUAUCUGCUCAAGCCCAUUCCUUACCUGCUUUCAACACUUGUAAA-GUUACGCAGGUAGGAAACUUGGGCAC-------------------GGAUUCAUUU ----.(((((((((..(((.....)))....((((((((((.((((((....(((.........-)))..)))))))))))...)))))))-------------------))).)))).. ( -32.60) >DroEre_CAF1 162718 96 - 1 ----UAUUAUACGCCUGUACUAUCUGCUCAAGCCCAUUCCUUACCAGCUUUCAACACUUGUAAA-GUUACGCAGGUAGGAAACUUGGGCAC-------------------GGAUUCAUUU ----.................(((((.....((((((((((.(((((((((((.....)).)))-))).....))))))))...))))).)-------------------))))...... ( -20.30) >DroYak_CAF1 167023 96 - 1 ----UAUGGUCCUCCUGCACUAUCUGCUCAAGCACAUUCCUUAGCUGCUUUCAACACUUGUAAA-GUUACGCAGGUAGGAAACUUGGACAC-------------------GGAUUCAUUU ----....((((.......((((((((...(((..........)))(((((((.....)).)))-))...)))))))).......))))..-------------------.......... ( -21.94) >consensus ____UAUGGUCCGCCUGCACUAUCUGCUCAAGCCCAUUCCUUACCUGCUUUCAACACUUGUAAA_GUUACGCAGGUAGGAAACUUGGGCAC___________________GGAUUCAUUU ........................((((((((....(((((.((((((......................))))))))))).)))))))).............................. (-12.13 = -14.72 + 2.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:50 2006