
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,747,812 – 13,748,039 |
| Length | 227 |
| Max. P | 0.999798 |

| Location | 13,747,812 – 13,747,924 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 90.89 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -30.46 |
| Energy contribution | -30.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13747812 112 - 23771897 CACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAGCAUAGACUAACUGAACGAAAGCUGAAGUUGUCCCGCCUCCAAAGAGAAGGCGGAAGUUGGCCA ...(((((...(((((.....)))))...)))))...((((((..........(((.((((..((....).)..)))))))((((((.(.....).))))))..)))))).. ( -37.80) >DroSec_CAF1 154555 99 - 1 CACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAG-------------AACGAAAGCUGAAAUCGACCCGCCUCCAAAGCGAAGGCGGAAGUUGGCCA ...(((((...(((((.....)))))...)))))...((((((.(((..-------------..(....))))........((((((.(.....).))))))..)))))).. ( -32.90) >DroSim_CAF1 166198 112 - 1 CACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAGCAUAGUCUAACUGAACGAAAGCUGAAGUCGACCCGCCUCCAAAGCGAAGGCGGAAGUUGGCCA ......(((((....)))))(((((((.(((.....(((....))).))).)))))))......(....).....((((((((((((.(.....).))))))..)))))).. ( -35.60) >DroEre_CAF1 161416 112 - 1 CACUCCGGAGGAGUCCUUCCUGGACUGAGUUAUAAAUGCCAACGCAGAGCAUAGUCUAACUUAACGAAAGCUGAAGUCGUCCCGCCUCCAAAGCGAAGGCGGAAGUUGGCCA ((.((((((((....))))).))).))..........((((((...............((((..(....)...))))....((((((.(.....).))))))..)))))).. ( -33.60) >DroYak_CAF1 165693 112 - 1 CACUGUGGAGGAGUCCUUCCUGGACUGAGUUAUAAAUGCCAACGCAGAGCAUAGUCUAACUGAACGAAAGCCGAAGUCGUCCCGCCACCAAAGCGAUGGCGGAAGUUGGCCA ......(((((....)))))(((((((.(((.....(((....))).))).)))))))...........(((((.......((((((.(.....).))))))...))))).. ( -31.90) >consensus CACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAGCAUAGUCUAACUGAACGAAAGCUGAAGUCGUCCCGCCUCCAAAGCGAAGGCGGAAGUUGGCCA ...(((((...(((((.....)))))...)))))...((((((.....................(....)...........((((((.(.....).))))))..)))))).. (-30.46 = -30.30 + -0.16)

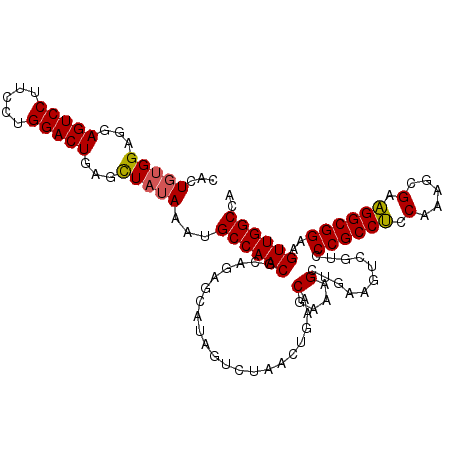

| Location | 13,747,851 – 13,747,964 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13747851 113 - 23771897 CGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAGCAUAGACUAACUGAACGAAAGCUG ...........(((.........((((.....((((((........))))))((((.....)))))))).......(((....))))))...............(....)... ( -21.40) >DroSec_CAF1 154594 100 - 1 CGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAG-------------AACGAAAGCUG ...........(((.........((((.....((((((........))))))((((.....)))))))).......(((....))))))-------------..(....)... ( -22.10) >DroSim_CAF1 166237 113 - 1 CGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAGCAUAGUCUAACUGAACGAAAGCUG ..................................(((((((.....)))))))((.(((.(((((((.(((.....(((....))).))).)))))))...))).))...... ( -24.50) >DroEre_CAF1 161455 113 - 1 CGACCAACCCACUCAUACACUCCCUCAACACUCCCUCCUCCACUCCGGAGGAGUCCUUCCUGGACUGAGUUAUAAAUGCCAACGCAGAGCAUAGUCUAACUUAACGAAAGCUG .(((......(((((....................((((((.....))))))((((.....))))))))).....(((((......).)))).)))........(....)... ( -25.50) >DroYak_CAF1 165732 106 - 1 CGACCAACCCACUCAUACACCCCC-------UCCCUCCUUCACUGUGGAGGAGUCCUUCCUGGACUGAGUUAUAAAUGCCAACGCAGAGCAUAGUCUAACUGAACGAAAGCCG .(((......((((..........-------..(((((........)))))(((((.....))))))))).....(((((......).)))).)))........(....)... ( -25.30) >consensus CGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAUAAAUGCCAACGCAGAGCAUAGUCUAACUGAACGAAAGCUG ...........(((.........(((.......(((((........)))))(((((.....)))))))).......(((....))))))...............(....)... (-19.62 = -19.78 + 0.16)


| Location | 13,747,891 – 13,747,999 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.52 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -27.28 |
| Energy contribution | -26.64 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13747891 108 + 23771897 AUAGCUCAGUCCAGGAAGGACUCCUCCACAGUGGAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGGGCAAU ...(((((((((.....)))))((((.(((.(.........).))).))))..(((.((...((.((..(((((......)))))..)).)).)))))...))))... ( -31.50) >DroSec_CAF1 154621 108 + 1 AUAGCUCAGUCCAGGAAGGACUCCUCCACAGUGGAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGGGCAAU ...(((((((((.....)))))((((.(((.(.........).))).))))..(((.((...((.((..(((((......)))))..)).)).)))))...))))... ( -31.50) >DroSim_CAF1 166277 108 + 1 AUAGCUCAGUCCAGGAAGGACUCCUCCACAGUGGAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAAGGCAAU ...(((((((((.....)))))((((.(((.(.........).))).))))))))...(((....((..(((((......)))))..)).....)))........... ( -29.80) >DroEre_CAF1 161495 108 + 1 AUAACUCAGUCCAGGAAGGACUCCUCCGGAGUGGAGGAGGGAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGGGCAAU ....(((..(((....((.((((((((........))).))))).))...)))(((.((...((.((..(((((......)))))..)).)).))))).)))...... ( -32.30) >DroYak_CAF1 165772 101 + 1 AUAACUCAGUCCAGGAAGGACUCCUCCACAGUGAAGGAGGGA-------GGGGGUGUAUGAGUGGGUUGGUCGUCUGAGAGUGACUGGCAUAAAUCACAGAGGGCAAU .(((((((((((.....))))((((((........)))))).-------.............)))))))(((.((((.(((((.....)))...)).)))).)))... ( -30.70) >consensus AUAGCUCAGUCCAGGAAGGACUCCUCCACAGUGGAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGGGCAAU .(((((((((((.....))))(((((..........))))).....................)))))))(((.((((.(((((.....)))...)).)))).)))... (-27.28 = -26.64 + -0.64)

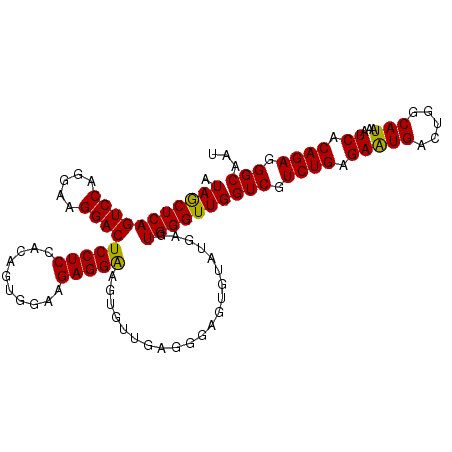

| Location | 13,747,891 – 13,747,999 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.52 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -22.42 |
| Energy contribution | -21.86 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13747891 108 - 23771897 AUUGCCCUCUGUGAUUUAUGCCAGUCAUUCUCAGACGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAU .(((..((((((((((......)))))....)))).)..)))................((((.....((((((........))))))((((.....)))))))).... ( -21.20) >DroSec_CAF1 154621 108 - 1 AUUGCCCUCUGUGAUUUAUGCCAGUCAUUCUCAGACGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAU .(((..((((((((((......)))))....)))).)..)))................((((.....((((((........))))))((((.....)))))))).... ( -21.20) >DroSim_CAF1 166277 108 - 1 AUUGCCUUCUGUGAUUUAUGCCAGUCAUUCUCAGACGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAU ..........((((((......)))))).(((((...................................(((((((.....)))))))(((.....)))))))).... ( -20.90) >DroEre_CAF1 161495 108 - 1 AUUGCCCUCUGUGAUUUAUGCCAGUCAUUCUCAGACGACCAACCCACUCAUACACUCCCUCAACACUCCCUCCUCCACUCCGGAGGAGUCCUUCCUGGACUGAGUUAU .(((..((((((((((......)))))....)))).)..)))...(((((....................((((((.....))))))((((.....)))))))))... ( -24.20) >DroYak_CAF1 165772 101 - 1 AUUGCCCUCUGUGAUUUAUGCCAGUCACUCUCAGACGACCAACCCACUCAUACACCCCC-------UCCCUCCUUCACUGUGGAGGAGUCCUUCCUGGACUGAGUUAU ..........((((((......)))))).................((((..........-------..(((((........)))))(((((.....)))))))))... ( -24.20) >consensus AUUGCCCUCUGUGAUUUAUGCCAGUCAUUCUCAGACGACCAACCCACUCAUACACUCCCUCAACACUUCCUCUUCCACUGUGGAGGAGUCCUUCCUGGACUGAGCUAU ..........((((((......)))))).(((((...................................(((((((.....)))))))(((.....)))))))).... (-22.42 = -21.86 + -0.56)

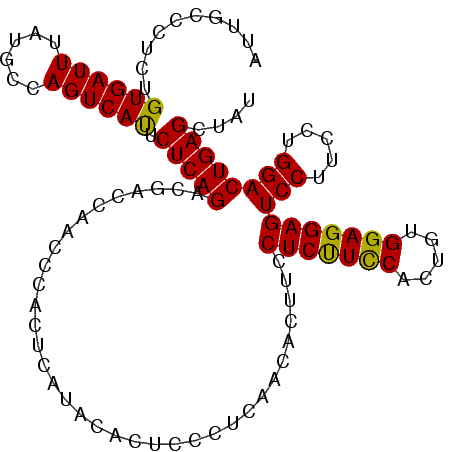

| Location | 13,747,924 – 13,748,039 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -22.81 |
| Energy contribution | -23.87 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13747924 115 + 23771897 GAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGG-----GCAAUAUGCAACACUAUUUCCAGUACGGCACUGCAAAUACUCCUC ...(((((.(((((...(.(((((...(.(((((..(((((......)))))..))..............-----((.....))..........))).)...))))).).)))))))))) ( -35.50) >DroSec_CAF1 154654 115 + 1 GAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGG-----GCAAUAUGCAACACUAUUUCCAGUGCGGCACUGCAAAUACUCCUC ...(((((.(((((...(.(((((...(.(((((..(((((......)))))..))..............-----((.....))..........))).)...))))).).)))))))))) ( -34.40) >DroSim_CAF1 166310 115 + 1 GAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAAG-----GCAAUAUGCAACACUAUUUCCAGUGCGGCACUGCAAAUACUCCUC ...(((((.(((((...(.(((((...(.(((((..(((((......)))))..))..............-----((.....))..........))).)...))))).).)))))))))) ( -34.40) >DroEre_CAF1 161528 115 + 1 GAGGAGGGAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGG-----GCAAUAUGCAACACUAUUUCCAGAGCGGCACUGCAAAUACUCCUC (((((((.(((((((..((((......((((.((..(((((......)))))..))..............-----((.....))..))))..))))....))))))).).....)))))) ( -36.50) >DroYak_CAF1 165805 108 + 1 AAGGAGGGA-------GGGGGUGUAUGAGUGGGUUGGUCGUCUGAGAGUGACUGGCAUAAAUCACAGAGG-----GCAAUAUGCAACACUAUUUCCAGUGCGGCACUGCAAAUACUCCUG .((((((((-------((.(((((.(((....((..((((........))))..)).....)))......-----((.....)).))))).)))))..((((....))))....))))). ( -33.30) >DroPer_CAF1 158330 101 + 1 -------CA-------CAAUGUG----CUUGGGUGGGUGGUGC-CGAGUGACUGGCAUAAAUCACUGGGCGCCACGCAGUAUGCAACAUUAUUUUUAGUUCAGCAUUGUAAAUACUCGCU -------.(-------(((((((----(...(.((.(((((((-(.(((((.(......).))))).)))))))).)).)..))...((((....))))...)))))))........... ( -36.40) >consensus GAAGAGGAAGUGUUGAGGGAGUGUAUGAGUGGGUUGGUCGUCUGAGAAUGACUGGCAUAAAUCACAGAGG_____GCAAUAUGCAACACUAUUUCCAGUGCGGCACUGCAAAUACUCCUC ................((((((((....((((((..(((((......)))))..)).....))))..........(((...(((..((((......))))..))).)))..)))))))). (-22.81 = -23.87 + 1.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:47 2006