| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,744,113 – 13,744,205 |
| Length | 92 |
| Max. P | 0.700155 |

| Location | 13,744,113 – 13,744,205 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.99 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
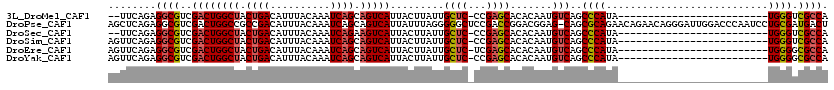
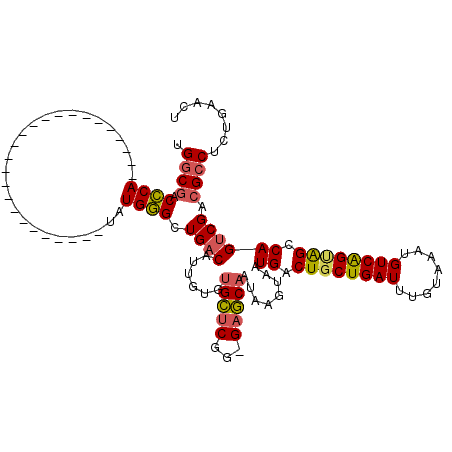
>3L_DroMel_CAF1 13744113 92 + 23771897 --UUCAGAGGCGUCGACUGGCUACUGACAUUUACAAAUCAGCAGUCAUUACUUAUUGCUC-CCGAGCACACAAUGUCAGCCCAUA-------------------------UGGGUCGCCA --......((((..((((((((.((((..........)))).)))))........((((.-...))))......))).((((...-------------------------.)))))))). ( -26.40) >DroPse_CAF1 151947 119 + 1 AGCUCAGAGGCGUCGACUGGCCGCCGACAUUUACAAAUCAGCAGUCAUUAUUUAGGGGGCUCCGACCGGACGGAG-CAGCGCAGAACAGAACAGGGAUUGGACCCAAUCCUGCGAUGACU .(((....((((((....)).))))((..........)))))(((((((.....(.(.((((((......)))))-)..).)...........(((((((....)))))))..))))))) ( -36.10) >DroSec_CAF1 150870 92 + 1 --UUCAGAGGCGUCGACUGGCUACUGACAUUUACAAAUCAGAAGUCAUUACUUAUUGCUC-CCGAGCACACAAUGUCAGCCCAUA-------------------------UGGGUCGCCA --......((((..((((((((.((((..........)))).)))))........((((.-...))))......))).((((...-------------------------.)))))))). ( -26.40) >DroSim_CAF1 162005 94 + 1 AGUUCAGAGGCGUCGACUGGCUACUGACAUUUACAAAUCAGCAGUCAUUACUUAUUGCUC-CCGAGCACACAAUGUCAGCCCAUA-------------------------UGGGUCGCCA ........((((..((((((((.((((..........)))).)))))........((((.-...))))......))).((((...-------------------------.)))))))). ( -26.40) >DroEre_CAF1 157763 94 + 1 AGUUCAGAGGCGUCGACUGGCUACUGACAUUUACAAAUCAGCAGUCAUUACUUAUUGCUC-UCGAGCACACAAUGUCAGCCCAUA-------------------------UGGGGCGCCA ........(((...((((((((.((((..........)))).)))))........((((.-...))))......))).((((...-------------------------..))))))). ( -27.80) >DroYak_CAF1 161918 94 + 1 AGUUCAGAGGCGUCGACUGGCUACUGACAUUUACAAAUCAGCAGUCAUUACUUAUUGCUC-CCGAGCACACAAUGUCAGCCCAUA-------------------------UGGGGCGCCA ........(((...((((((((.((((..........)))).)))))........((((.-...))))......))).((((...-------------------------..))))))). ( -27.80) >consensus AGUUCAGAGGCGUCGACUGGCUACUGACAUUUACAAAUCAGCAGUCAUUACUUAUUGCUC_CCGAGCACACAAUGUCAGCCCAUA_________________________UGGGUCGCCA ........((((..((((((((.((((..........)))).))))).........((((...)))).......)))..((((...........................)))).)))). (-19.94 = -20.08 + 0.14)
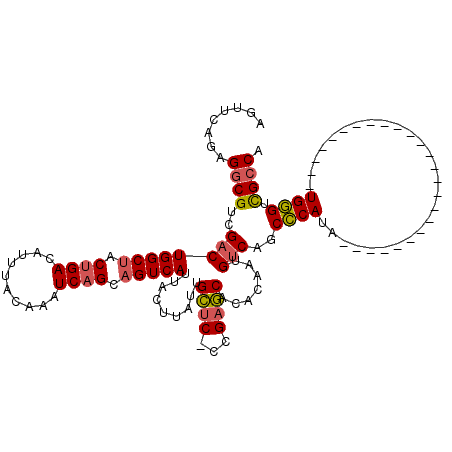
| Location | 13,744,113 – 13,744,205 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.99 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13744113 92 - 23771897 UGGCGACCCA-------------------------UAUGGGCUGACAUUGUGUGCUCGG-GAGCAAUAAGUAAUGACUGCUGAUUUGUAAAUGUCAGUAGCCAGUCGACGCCUCUGAA-- .((((.(((.-------------------------...))).(((((((...((((...-.))))...)))..((.((((((((........)))))))).)))))).))))......-- ( -30.30) >DroPse_CAF1 151947 119 - 1 AGUCAUCGCAGGAUUGGGUCCAAUCCCUGUUCUGUUCUGCGCUG-CUCCGUCCGGUCGGAGCCCCCUAAAUAAUGACUGCUGAUUUGUAAAUGUCGGCGGCCAGUCGACGCCUCUGAGCU ((((((((((((((.(((......)))......))))))))..(-(((((......))))))..........))))))((((((........))))))((((((.........))).))) ( -41.00) >DroSec_CAF1 150870 92 - 1 UGGCGACCCA-------------------------UAUGGGCUGACAUUGUGUGCUCGG-GAGCAAUAAGUAAUGACUUCUGAUUUGUAAAUGUCAGUAGCCAGUCGACGCCUCUGAA-- .((((.(((.-------------------------...)))((((((((......((((-(((((........)).)))))))......))))))))...........))))......-- ( -28.90) >DroSim_CAF1 162005 94 - 1 UGGCGACCCA-------------------------UAUGGGCUGACAUUGUGUGCUCGG-GAGCAAUAAGUAAUGACUGCUGAUUUGUAAAUGUCAGUAGCCAGUCGACGCCUCUGAACU .((((.(((.-------------------------...))).(((((((...((((...-.))))...)))..((.((((((((........)))))))).)))))).))))........ ( -30.30) >DroEre_CAF1 157763 94 - 1 UGGCGCCCCA-------------------------UAUGGGCUGACAUUGUGUGCUCGA-GAGCAAUAAGUAAUGACUGCUGAUUUGUAAAUGUCAGUAGCCAGUCGACGCCUCUGAACU .(((((((..-------------------------...))))(((((((...((((...-.))))...)))..((.((((((((........)))))))).))))))..)))........ ( -31.30) >DroYak_CAF1 161918 94 - 1 UGGCGCCCCA-------------------------UAUGGGCUGACAUUGUGUGCUCGG-GAGCAAUAAGUAAUGACUGCUGAUUUGUAAAUGUCAGUAGCCAGUCGACGCCUCUGAACU .(((((((..-------------------------...))))(((((((...((((...-.))))...)))..((.((((((((........)))))))).))))))..)))........ ( -31.30) >consensus UGGCGACCCA_________________________UAUGGGCUGACAUUGUGUGCUCGG_GAGCAAUAAGUAAUGACUGCUGAUUUGUAAAUGUCAGUAGCCAGUCGACGCCUCUGAACU .((((.((((...........................)))).((((......(((((...)))))........((.((((((((........)))))))).)))))).))))........ (-24.12 = -24.18 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:38 2006