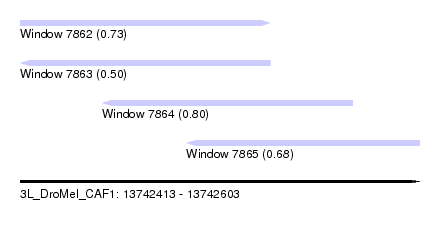
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,742,413 – 13,742,603 |
| Length | 190 |
| Max. P | 0.795330 |

| Location | 13,742,413 – 13,742,532 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -36.61 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13742413 119 + 23771897 UUGGCUUGAGUUGUUCAGCC-CCAAACCAUCAGGGCAGAGCAAGCGAGCCGGAGCAAAAAGAAAGAAGUCAGUUGGUGCAAUUCAAGCCAAUGUCAAAUCCAUUGCUGGACGACUGGCUU ((((((((..((((((.(((-(..........)))).)))))).)))))))).............(((((((((((.((.......))).........((((....)))))))))))))) ( -40.20) >DroSec_CAF1 149139 119 + 1 UUGGCUUGAGUUGUUCACCC-CCAAACCAUCAGGGCAGAGCAAGCGAGACGGAGCAAAAAGAAAGAAGUCAGUUGGUGCAAUUCAAGCCAAUGUCAAAUCCAUUGCUGGACGACUGGCUU (((((((((((((..(((((-((.........))).......(((..(((.................))).)))))))))))))))))))).((((..((((....))))....)))).. ( -33.43) >DroSim_CAF1 160277 119 + 1 UUGGCUUGAGUUGUUCAGCC-CCAAACCAUCAGGGCAGAGCAAGCGAGACGGAGCAAAAAGAAAGAAGUCAGUUGGUGCAAUUCAAGCCAAUGUCAAAUCCAUUGCUGGACGACUGGCUU ((((((((((((((((.(((-(..........)))).))))..(((.(((...((............))..)))..))))))))))))))).((((..((((....))))....)))).. ( -35.80) >DroEre_CAF1 155763 119 + 1 UUGGCUUGAGUUGUUCAGAC-CCAAGCCAUCAGGACAGAGCAAGCGAGCCGAAGCAAAAAGAAAGAAGUCAGUUGGUGCAAUUCAAGCCAAUGUCAAAUCCAUUGCUGGACGACUGGCUU ((((((((..((((((.(.(-(..........)).).)))))).)))))))).............(((((((((((.((.......))).........((((....)))))))))))))) ( -33.50) >DroYak_CAF1 160115 120 + 1 UUGGCUUGAGUUGUUCAGCCCCCAAACCAUCAGGGCAAAGCUGGCGAGCCGGAGCAGAAAGAAAGAAGUCAGUUGCUGCAAUUCAAGCCGAUGUCAAAUCCAUUGCUGGACGACUGGCUU ((((((((((((((...((((...........))))....((((....))))(((((...((......))..))))))))))))))))))).((((..((((....))))....)))).. ( -40.10) >consensus UUGGCUUGAGUUGUUCAGCC_CCAAACCAUCAGGGCAGAGCAAGCGAGCCGGAGCAAAAAGAAAGAAGUCAGUUGGUGCAAUUCAAGCCAAUGUCAAAUCCAUUGCUGGACGACUGGCUU (((((((((((((((((((.......((....)).........((........))................))))).)))))))))))))).((((..((((....))))....)))).. (-30.44 = -30.88 + 0.44)



| Location | 13,742,413 – 13,742,532 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13742413 119 - 23771897 AAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGGCUCGCUUGCUCUGCCCUGAUGGUUUGG-GGCUGAACAACUCAAGCCAA ......((((((((....)))))..))).(((((((((.(((.............................((......))((.((((..(....)..)-))).)).))).))))))))) ( -36.60) >DroSec_CAF1 149139 119 - 1 AAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGUCUCGCUUGCUCUGCCCUGAUGGUUUGG-GGGUGAACAACUCAAGCCAA ......((((((((....)))))..))).(((((((((.(((((((..(.((......))........(((((..((.......))...)))))...).-.))))..))).))))))))) ( -36.20) >DroSim_CAF1 160277 119 - 1 AAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGUCUCGCUUGCUCUGCCCUGAUGGUUUGG-GGCUGAACAACUCAAGCCAA ......((((((((....)))))..))).(((((((((...(((......(((.................))).....)))((.((((..(....)..)-))).)).....))))))))) ( -36.13) >DroEre_CAF1 155763 119 - 1 AAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUUCGGCUCGCUUGCUCUGUCCUGAUGGCUUGG-GUCUGAACAACUCAAGCCAA ((((((((((((((....)))))..)))..))))))..............(((............((....))...........)))....((((((((-((.......)))))))))). ( -36.30) >DroYak_CAF1 160115 120 - 1 AAGCCAGUCGUCCAGCAAUGGAUUUGACAUCGGCUUGAAUUGCAGCAACUGACUUCUUUCUUUCUGCUCCGGCUCGCCAGCUUUGCCCUGAUGGUUUGGGGGCUGAACAACUCAAGCCAA ......((((((((....)))))..)))...(((((((.(((((((...................(((..((....)))))....(((..(....)..)))))))..))).))))))).. ( -39.40) >consensus AAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGGCUCGCUUGCUCUGCCCUGAUGGUUUGG_GGCUGAACAACUCAAGCCAA ......((((((((....)))))..))).(((((((((.(((.......................((........))....((.((((..(....)..).))).)).))).))))))))) (-28.20 = -28.64 + 0.44)


| Location | 13,742,452 – 13,742,571 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.74 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13742452 119 - 23771897 -GCCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGGCUCGCUUG -(((((((....))))...)))(((..((.((.......(((((((((((((((....)))))..)))..)))))))....(((......((......))....))))).))..)))... ( -32.50) >DroSec_CAF1 149178 119 - 1 -GUCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGUCUCGCUUG -.............(((((((((................(((((((((((((((....)))))..)))..)))))))....(((......((......))....)))..))))))))).. ( -33.20) >DroSim_CAF1 160316 119 - 1 -GUCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGUCUCGCUUG -.............(((((((((................(((((((((((((((....)))))..)))..)))))))....(((......((......))....)))..))))))))).. ( -33.20) >DroEre_CAF1 155802 120 - 1 CACCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUUCGGCUCGCUUG ......((((..((((((..(((((((...)))))....(((((((((((((((....)))))..)))..)))))))....))..).)))))....)))).....((....))....... ( -33.80) >DroYak_CAF1 160155 120 - 1 AGCCCUGGAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUCGGCUUGAAUUGCAGCAACUGACUUCUUUCUUUCUGCUCCGGCUCGCCAG ((((..(((......((.(((......))).))......((((((.((((((((....)))))..)))...))))))....((((.((..((......)).)))))))))))))...... ( -33.30) >consensus _GCCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAUUGCACCAACUGACUUCUUUCUUUUUGCUCCGGCUCGCUUG ......((((..((((((..(((((((...)))))....(((((((((((((((....)))))..)))..)))))))....))..).)))))....)))).....((........))... (-29.70 = -29.74 + 0.04)


| Location | 13,742,492 – 13,742,603 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -24.24 |
| Energy contribution | -23.83 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13742492 111 - 23771897 AUGCCCUGCCACACCGACACCCGCUCGUUGCC---------GCCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAU ...........((..(((((..((.....)).---------(((((((....))))...))))))))..))........(((((((((((((((....)))))..)))..)))))))... ( -32.90) >DroSec_CAF1 149218 111 - 1 AUGCCCUGCCACACCGACACCCUCUCGUUGCC---------GUCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAU ...........((..(((((((((.((....)---------)..((((....)))).)))).)))))..))........(((((((((((((((....)))))..)))..)))))))... ( -33.30) >DroSim_CAF1 160356 111 - 1 AUGCCCUGCCACACCGACACCCCCUCGUUGCC---------GUCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAU ..........(((((((.......)))..(((---------.((((((....)))).))))))))).............(((((((((((((((....)))))..)))..)))))))... ( -32.50) >DroEre_CAF1 155842 120 - 1 CUGCCCUGCCACACCGACACCCCCUCGUCGCCCCUCGUUCCACCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAU ..........((((((((........))))..(((((.......((((....))))))))).)))).............(((((((((((((((....)))))..)))..)))))))... ( -32.31) >DroYak_CAF1 160195 117 - 1 GUACCCCGCCACACUGACACCCUCUCUUCGCCCAGG---AAGCCCUGGAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUCGGCUUGAAU ...........((..(((((((((.((....(((((---....)))))......)).)))).)))))..))........((((((.((((((((....)))))..)))...))))))... ( -39.70) >DroPer_CAF1 150598 105 - 1 -UAUCCGGCUU---CGAU----CCUCGUCCCCCUGG---C-A-UCUGGCGAAUCAGUGAAGCGUGUUUUUGGCAUUUAACAAGCCGGUUGUCCAGCAAUGGGUUCGAAAUCAUCCUAA-- -.....(..((---(((.----((.(((....((((---(-(-.(((((.......((((((.........)).))))....))))).)).))))..))))).)))))..).......-- ( -27.70) >consensus AUGCCCUGCCACACCGACACCCCCUCGUCGCC_________GCCCUGAAAAAUCAGUGAGGCGUGUUCCUGACAUUUAACAAGCCAGUCGUCCAGCAAUGGAUUUGACAUUGGCUUGAAU .......(((....((((........))))..............((((....))))...)))(((((...)))))....(((((((((((((((....))))..))))..)))))))... (-24.24 = -23.83 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:34 2006