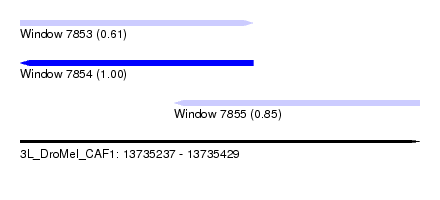
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,735,237 – 13,735,429 |
| Length | 192 |
| Max. P | 0.999693 |

| Location | 13,735,237 – 13,735,349 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -16.40 |
| Consensus MFE | -13.54 |
| Energy contribution | -13.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13735237 112 + 23771897 AAGUUAUGAAUCAAAAAUAAUAUAGAAAAA-AA---GAUGAGGCGUAGAAAA-GAGGCCAGAUAGAACGG-GGGAAACUUUUUAUGAUUUCAAUAAAGUUGUUAACUUUAUUAAAAUU ((((((((((....................-..---.....(((.(......-.).)))..........(-((....))))))))))))).((((((((.....))))))))...... ( -16.10) >DroSec_CAF1 142107 112 + 1 AAGUUAUGAAUCAAAAAUAAUAUAGAAAAA-AA---GAUGAGGCGUAGAAAA-GAGACCAGAUAGAACGG-GGGAAACUUUUUAUGAUUUCAAUAAAGUUGUUAACUUUAUUAAAAUU ..(((((.........))))).........-..---...((((((((((((.-....((.(......).)-)(....))))))))).))))((((((((.....))))))))...... ( -13.70) >DroSim_CAF1 152852 111 + 1 AAGUUAUGAAUCAAAAAUAAUAUA-AAAAA-AA---GAUGAGGCGUAGAAAA-GAGGCCAGAUAGAACGG-GGGAAACUUUUUAUGAUUUCAAUAAAGUUGUUAACUUUAUUAAAAUU ((((((((((..............-.....-..---.....(((.(......-.).)))..........(-((....))))))))))))).((((((((.....))))))))...... ( -16.10) >DroEre_CAF1 148694 113 + 1 AAGUUAUGAAUCAAAAAUAAUGUAGAAAAAAAA---AAUGAGGCGAAGAAAA-GAGGCCAGAUAGAACGG-GGGAAACUUUUUAUGAUUUCAAUAAAGUUGUUAACUUUAUUAAAAUU ((((((((((.......................---.....(((........-...)))..........(-((....))))))))))))).((((((((.....))))))))...... ( -15.40) >DroYak_CAF1 152767 118 + 1 AAGUUAUGAAUCAAAAAUAAUAUAGAAAAAAAAGAAGAUGAGGCCAAGAAAACGGGGCCAGAUAGAAGGGGGGGAAACUUUUUAUGAUUUCAAUAAAGUUGUUAACUUUAUUAAAAUU .......((((((............................((((..(....)..))))........((((((....)))))).)))))).((((((((.....))))))))...... ( -20.70) >consensus AAGUUAUGAAUCAAAAAUAAUAUAGAAAAA_AA___GAUGAGGCGUAGAAAA_GAGGCCAGAUAGAACGG_GGGAAACUUUUUAUGAUUUCAAUAAAGUUGUUAACUUUAUUAAAAUU ((((((((((...............................(((............)))............((....)).)))))))))).((((((((.....))))))))...... (-13.54 = -13.74 + 0.20)

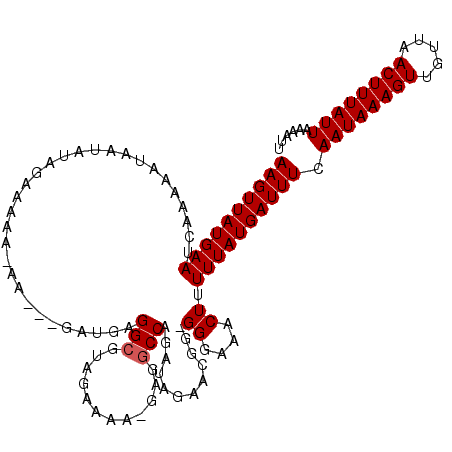

| Location | 13,735,237 – 13,735,349 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -12.04 |
| Consensus MFE | -11.52 |
| Energy contribution | -11.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13735237 112 - 23771897 AAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAAAAAGUUUCCC-CCGUUCUAUCUGGCCUC-UUUUCUACGCCUCAUC---UU-UUUUUCUAUAUUAUUUUUGAUUCAUAACUU .(((((((((((((.....)))))))))))))...............-...........(((...-........))).....---..-.............................. ( -11.40) >DroSec_CAF1 142107 112 - 1 AAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAAAAAGUUUCCC-CCGUUCUAUCUGGUCUC-UUUUCUACGCCUCAUC---UU-UUUUUCUAUAUUAUUUUUGAUUCAUAACUU .(((((((((((((.....)))))))))))))....(((((...((.-...........))...)-))))............---..-.............................. ( -10.70) >DroSim_CAF1 152852 111 - 1 AAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAAAAAGUUUCCC-CCGUUCUAUCUGGCCUC-UUUUCUACGCCUCAUC---UU-UUUUU-UAUAUUAUUUUUGAUUCAUAACUU .(((((((((((((.....))))))))))))).((((((((......-..(......).(((...-........))).....---..-)))))-)))..................... ( -11.80) >DroEre_CAF1 148694 113 - 1 AAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAAAAAGUUUCCC-CCGUUCUAUCUGGCCUC-UUUUCUUCGCCUCAUU---UUUUUUUUCUACAUUAUUUUUGAUUCAUAACUU .(((((((((((((.....)))))))))))))...............-...........(((...-........))).....---................................. ( -11.40) >DroYak_CAF1 152767 118 - 1 AAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAAAAAGUUUCCCCCCCUUCUAUCUGGCCCCGUUUUCUUGGCCUCAUCUUCUUUUUUUUCUAUAUUAUUUUUGAUUCAUAACUU .(((((((((((((.....)))))))))))))...........................((((..(....)..))))......................................... ( -14.90) >consensus AAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAAAAAGUUUCCC_CCGUUCUAUCUGGCCUC_UUUUCUACGCCUCAUC___UU_UUUUUCUAUAUUAUUUUUGAUUCAUAACUU .(((((((((((((.....)))))))))))))...........................(((............)))......................................... (-11.52 = -11.36 + -0.16)

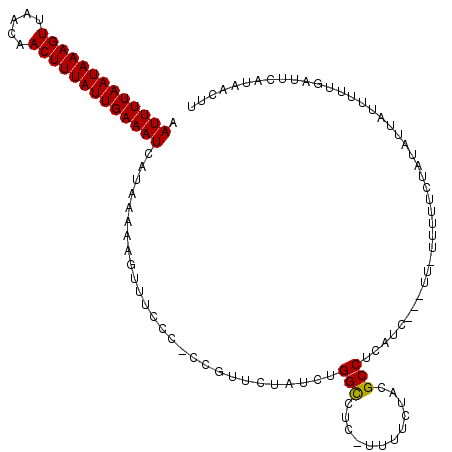
| Location | 13,735,311 – 13,735,429 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.43 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13735311 118 - 23771897 GUCGUAGUCGGCGAAAGUAUUUUCUCAUAGUAAUUCUAUGCUCGGCUGGCUUUUAGACAACUUUUCUUGAGCUGAGCAAAAAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAA--A ((((....))))..........................(((((((((.(.....((....)).....).)))))))))...(((((((((((((.....))))))))))))).....--. ( -27.60) >DroPse_CAF1 136265 112 - 1 GUCGU------CGAAAGUAUUUUCUCAUAGUAAUUCUCUGCUCGGCUGGCUUUUGCACAACUUUUCUU--GCUGAGCAAAAAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCGUAAAAA .....------(((........................((((((((((((....)).)).........--))))))))....((((((((((((.....)))))))))))))))...... ( -23.50) >DroSim_CAF1 152925 118 - 1 GUCGUAGUCGGCGAAAGUAUUUUCUCAUAGUAAUUCUAUGCUCGGCUGGCUUUUAGACAACUUUUCUUGAGCUGAGCAAAAAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAA--A ((((....))))..........................(((((((((.(.....((....)).....).)))))))))...(((((((((((((.....))))))))))))).....--. ( -27.60) >DroEre_CAF1 148769 118 - 1 GUCGUAGUCGGCGAAAGUAUUUUCUCAUAGUAAUUCUAUGCUCGGCUGGCUUUUAGACAACUUUUCUUGAGCUGAGCAAAAAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAA--A ((((....))))..........................(((((((((.(.....((....)).....).)))))))))...(((((((((((((.....))))))))))))).....--. ( -27.60) >DroYak_CAF1 152847 118 - 1 GUCGUAGUCGGCGAAAGUAUUUUUUCAUAGUAAUUCUAUGCUCGGCUGGCUUUUAGACAACUUUUCUUGAGCUGAGCAAAAAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAA--A ((((....))))(((((....)))))............(((((((((.(.....((....)).....).)))))))))...(((((((((((((.....))))))))))))).....--. ( -29.90) >DroPer_CAF1 137592 112 - 1 GUCGU------CGAAAGUAUUUUCUCAUAGUAAUUCUCUGCUCGGCUGGCUUUUGCACAACUUUUCUU--GCUGAGCAAAAAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCGUAAAAA .....------(((........................((((((((((((....)).)).........--))))))))....((((((((((((.....)))))))))))))))...... ( -23.50) >consensus GUCGUAGUCGGCGAAAGUAUUUUCUCAUAGUAAUUCUAUGCUCGGCUGGCUUUUAGACAACUUUUCUUGAGCUGAGCAAAAAUUUUAAUAAAGUUAACAACUUUAUUGAAAUCAUAA__A .((((.....))))........................(((((((((......................)))))))))...(((((((((((((.....)))))))))))))........ (-22.25 = -22.92 + 0.67)

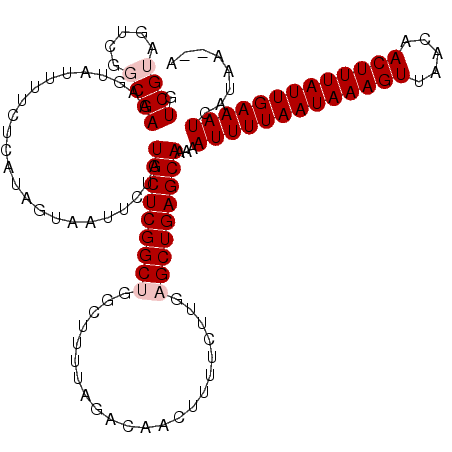
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:24 2006