| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,732,710 – 13,732,809 |
| Length | 99 |
| Max. P | 0.589637 |

| Location | 13,732,710 – 13,732,809 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -22.35 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13732710 99 + 23771897 AGUGGCAUGCCAGGAGCACAGGACU---------CGCAGGACACGCUGGGACAGCUGUCAUUAAGUGCAGG----AGCAAGCAUUUGAAUAAGCUU--ACUCAUGCUACAGUAU .((((((((....((((........---------.....((((.((((...)))))))).((((((((...----.....))))))))....))))--...))))))))..... ( -33.50) >DroSec_CAF1 139625 90 + 1 AGUGGCAUGCCAGGAGCACAG------------------GACACGCUGGGACAGCUGUCAUUAAGUGCAGG----AGCAAGCAUUUGAAUAAGCUU--ACUCAUGCUCCAGUAU .(..(((((....((((....------------------((((.((((...)))))))).((((((((...----.....))))))))....))))--...)))))..)..... ( -29.80) >DroSim_CAF1 150355 99 + 1 AGUGGCAUGCCAGGAGCACAGGACU---------CGCAGGACACGCUGGGACAGCUGUCAUUAAGUGCAGG----AGCAAGCAUUUGAAUAAGCUU--ACUCAUGCUCCAGUAU ..(((....)))((((((..((...---------.((..((((.((((...)))))))).((((((((...----.....))))))))....))..--.))..))))))..... ( -31.70) >DroEre_CAF1 146125 99 + 1 AGUGGCAUGGAAGGAGCACAGGACU---------CGCAGGACACGCCGGGACAGCUGUCAUUAAAUGCAGG----AGCAAGCAUUUGAAUAAGCUU--GCUCAUGCUCCAGUAU ............((((((.....((---------.(((.((((.((.......))))))......)))))(----(((((((..........))))--)))).))))))..... ( -32.00) >DroYak_CAF1 150163 108 + 1 AGUGGCAUGCCAGGAGCGCAGGACUCGCAGGACUCGCAGGACACGCUGGGACAGCUGUCAUUAAAUGCAGG----AGCAAGCAUUUGAAUAAGCUU--ACUCACGCUCCACUUU ..(((....)))((((((.((((.((....)).))((..((((.((((...)))))))).((((((((...----.....))))))))....))..--.))..))))))..... ( -33.30) >DroPer_CAF1 134797 107 + 1 AGUGGCAUGCCAC-AGGAUAGGAUG--CAG----AGCAGGACACGCAGGGACAGAUGUCAUUAAAUGCAGAAGGCAGCAAGCAUUUGAAUAAGUCAGAGCUUUUCCUCCCGGGC .((((....))))-.((..((((..--.((----(((..(((.......(((....))).((((((((............))))))))....)))...))))))))).)).... ( -28.80) >consensus AGUGGCAUGCCAGGAGCACAGGACU_________CGCAGGACACGCUGGGACAGCUGUCAUUAAAUGCAGG____AGCAAGCAUUUGAAUAAGCUU__ACUCAUGCUCCAGUAU ..(((....)))((((((..((.................((((.((((...)))))))).((((((((............))))))))...........))..))))))..... (-22.35 = -23.05 + 0.70)

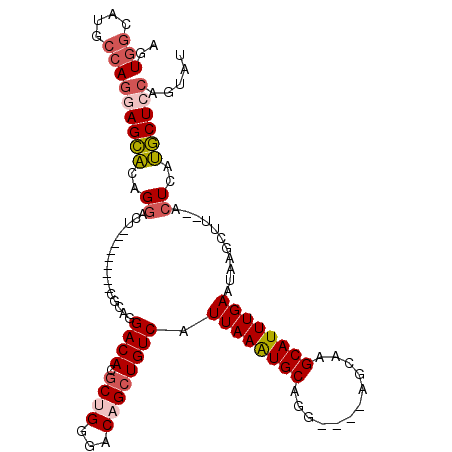

| Location | 13,732,710 – 13,732,809 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -18.33 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13732710 99 - 23771897 AUACUGUAGCAUGAGU--AAGCUUAUUCAAAUGCUUGCU----CCUGCACUUAAUGACAGCUGUCCCAGCGUGUCCUGCG---------AGUCCUGUGCUCCUGGCAUGCCACU ........(((.((((--((((..........)))))))----).))).......(((....)))...(((((((..(.(---------(((.....))))).))))))).... ( -30.70) >DroSec_CAF1 139625 90 - 1 AUACUGGAGCAUGAGU--AAGCUUAUUCAAAUGCUUGCU----CCUGCACUUAAUGACAGCUGUCCCAGCGUGUC------------------CUGUGCUCCUGGCAUGCCACU .....(((((((((((--((((..........)))))))----)...........((((((((...)))).))))------------------..)))))))(((....))).. ( -32.40) >DroSim_CAF1 150355 99 - 1 AUACUGGAGCAUGAGU--AAGCUUAUUCAAAUGCUUGCU----CCUGCACUUAAUGACAGCUGUCCCAGCGUGUCCUGCG---------AGUCCUGUGCUCCUGGCAUGCCACU .....(((((((((((--((((..........)))))))----)..(.((((.(.((((((((...)))).)))).)..)---------))).).)))))))(((....))).. ( -34.10) >DroEre_CAF1 146125 99 - 1 AUACUGGAGCAUGAGC--AAGCUUAUUCAAAUGCUUGCU----CCUGCAUUUAAUGACAGCUGUCCCGGCGUGUCCUGCG---------AGUCCUGUGCUCCUUCCAUGCCACU .....((.(((.((((--((((..........)))))))----).))).......(((....)))))((((((....(.(---------(((.....)))))...))))))... ( -34.10) >DroYak_CAF1 150163 108 - 1 AAAGUGGAGCGUGAGU--AAGCUUAUUCAAAUGCUUGCU----CCUGCAUUUAAUGACAGCUGUCCCAGCGUGUCCUGCGAGUCCUGCGAGUCCUGCGCUCCUGGCAUGCCACU ..(((((.(((.((((--((((..........)))))))----).))).......((((((((...)))).)))).(((.((....(((.......)))..)).)))..))))) ( -36.70) >DroPer_CAF1 134797 107 - 1 GCCCGGGAGGAAAAGCUCUGACUUAUUCAAAUGCUUGCUGCCUUCUGCAUUUAAUGACAUCUGUCCCUGCGUGUCCUGCU----CUG--CAUCCUAUCCU-GUGGCAUGCCACU (((((((((((...((...((((((((.((((((............))))))))))).....)))...))...)))(((.----..)--)).....))))-).)))........ ( -27.40) >consensus AUACUGGAGCAUGAGU__AAGCUUAUUCAAAUGCUUGCU____CCUGCACUUAAUGACAGCUGUCCCAGCGUGUCCUGCG_________AGUCCUGUGCUCCUGGCAUGCCACU .....((((((((((......)))........((.(((........)))......((((((((...)))).))))..))................)))))))(((....))).. (-18.33 = -19.20 + 0.87)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:21 2006