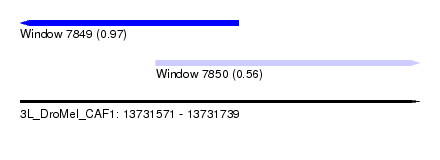
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,731,571 – 13,731,739 |
| Length | 168 |
| Max. P | 0.974123 |

| Location | 13,731,571 – 13,731,663 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -18.71 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
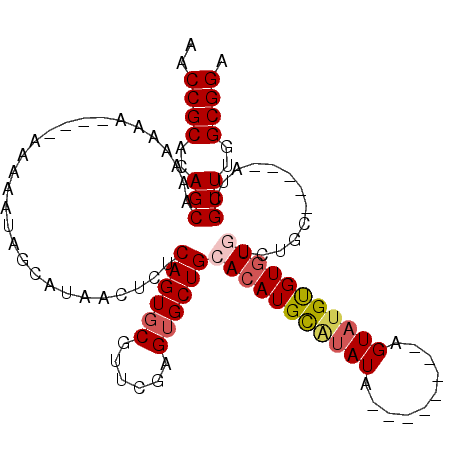
>3L_DroMel_CAF1 13731571 92 - 23771897 AACCGCACAGCAAAAAAAAAAAAA---AAGCAUAACUCUCAGUGCGUUCGAGUGCUGCACAUGUUUAUA--------GGUAUGUGUGU----GC-----AUCGUUUGGCGGA ..((((.(((..............---..((((........))))...(((.(((.((((((((.....--------...))))))))----))-----)))).))))))). ( -24.70) >DroSec_CAF1 138488 99 - 1 AACCGCACAGCAAAAAAAAAAAAAAAAUAGCAUAACUCUCAGUGCGUUCGAGUGCUGAACAUGUGUAUA--------GGUAUGUGUGUGUCUGC-----AUCGUUUGGCGGA ..((((.(((...................((((........))))...(((.(((.(((((((..((..--------..))..))))).)).))-----)))).))))))). ( -23.40) >DroEre_CAF1 144964 93 - 1 AACCGCACAGCAAAAA------AAAUAUGGCAUAACUCUCAGUGCAUUCGAGUGCUGCACAUGCAUAUA--------AGUAUGCGUGUGUCUGC-----AUCGUUUGGCGGA ..((((.(((......------.......((((........))))...(((.(((.(((((((((((..--------..)))))))))))..))-----)))).))))))). ( -32.80) >DroYak_CAF1 149026 108 - 1 AACCGCACAGCAAAAAAA----AAAAAUUGCAUAACUCUCAGUUCGUUCGAGUGCUGCACAUGCACAUAUAAGUACGAGUAUGUGUAUGUCUGCAUGGCAUCGUUUGGCGGA ..((((...((((.....----.....)))).............((..(((.((((((((((((((((((........))))))))))))..))..)))))))..)))))). ( -33.20) >consensus AACCGCACAGCAAAAAAA____AAAAAUAGCAUAACUCUCAGUGCGUUCGAGUGCUGCACAUGCAUAUA________AGUAUGUGUGUGUCUGC_____AUCGUUUGGCGGA ..((((..(((............................((((((......))))))(((((((((((..........))))))))))).............)))..)))). (-18.71 = -19.65 + 0.94)

| Location | 13,731,628 – 13,731,739 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
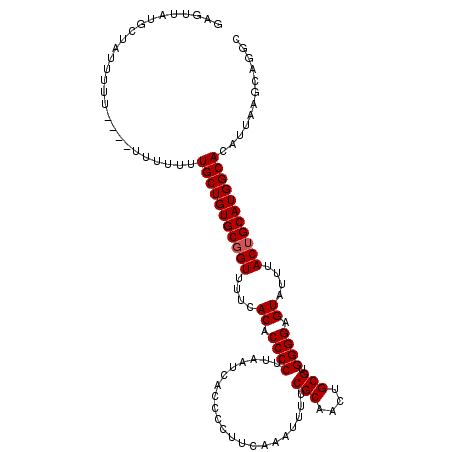
>3L_DroMel_CAF1 13731628 111 + 23771897 GAGUUAUGCUU---UUUUUUUUUUUUUGCUGUGCGGUUUUCACACCCCUUAAUCACCCCUUCAAAUUUUCGCAACUGCGUGGGGCGUAUUUACUGCAUGGCACAUUAAGCAGGC ......(((((---............(((((((((((....((.((((.....................(((....))).)))).))....)))))))))))....)))))... ( -28.84) >DroSec_CAF1 138549 114 + 1 GAGUUAUGCUAUUUUUUUUUUUUUUUUGCUGUGCGGUUUUCACACCCCUUAAUCACCCCUUCAAAUUUUCGCAACUGCGUGGGGAGUAUUUACGGCAUGGCACAUUAAGCAGGC ......((((................((((((((.((....((.((((.....................(((....))).)))).))....)).)))))))).....))))... ( -24.05) >DroEre_CAF1 145025 108 + 1 GAGUUAUGCCAUAUUU------UUUUUGCUGUGCGGUUUUCACACCCCUUAAUCACCCCUUCAAAUUUUCGCAACUGCGUGGGGAGUAUUUACUGCAUGGCACAUUAAGCAGGC .......(((...(((------..(.(((((((((((....((.((((.....................(((....))).)))).))....))))))))))).)..)))..))) ( -28.35) >DroYak_CAF1 149100 110 + 1 GAGUUAUGCAAUUUUU----UUUUUUUGCUGUGCGGUUUUCACACCCCUUAAUCACCCCUUGAAAUUUUCGCAACUGCGUGGGGAGUAUUUACUGCAUGGCACAUUAAGCAGGC ................----......(((((((((((....((.((((....(((.....)))......(((....))).)))).))....)))))))))))............ ( -27.80) >consensus GAGUUAUGCUAUUUUU____UUUUUUUGCUGUGCGGUUUUCACACCCCUUAAUCACCCCUUCAAAUUUUCGCAACUGCGUGGGGAGUAUUUACUGCAUGGCACAUUAAGCAGGC ..........................(((((((((((....((.((((.....................(((....))).)))).))....)))))))))))............ (-24.92 = -25.17 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:19 2006