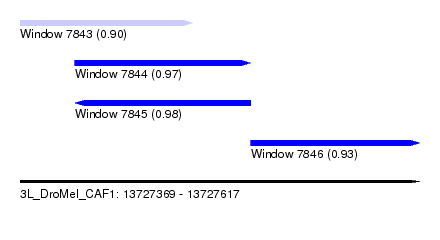
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,727,369 – 13,727,617 |
| Length | 248 |
| Max. P | 0.980937 |

| Location | 13,727,369 – 13,727,476 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -25.52 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13727369 107 + 23771897 GCAAUGAUAACCCUGCAAACAUGGCCCUGGCCCAAAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGGCAGCAGCAAAGGGCAGCUGGCA----- .....(((..((((((......(((....))).....((((........))))....))))))..)))..............((.((((.((.....)).)))).))----- ( -30.90) >DroSec_CAF1 134299 107 + 1 GCAAUGAUAACCCAGCAAACAUGGCCCUGGCCCAAAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA----- ...........(((((......(((....)))......(((........)))((((((((((....))))))..........((....)).....)))).)))))..----- ( -29.00) >DroSim_CAF1 145084 107 + 1 GCAAUGAUAACCCAGCAAACAUGGCCCUGGCCCAAAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA----- ...........(((((......(((....)))......(((........)))((((((((((....))))))..........((....)).....)))).)))))..----- ( -29.00) >DroEre_CAF1 140979 107 + 1 GCAAUGAUAACCCAGCAAACAUGGCCCUGGCCCAAAACGUUGAAAAUUAAACGGCCGGUAGGAGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA----- ...........(((((.......(((((((((......(((........)))))))((((((....))))))......................))))).)))))..----- ( -28.70) >DroYak_CAF1 144790 109 + 1 GCAAUGAUAACCCGGCAAACAUGGCCCUGGCCCAAAACGUUGAAAAUUAAACGCCCGGCAGGAGAGUCUACCAAAAUAUUUAUGACAGCAG---AGGGCAGCUGGCAAACUG ...........(((((......(((....)))......(((........)))((((((.(((....))).))..........((....)).---.)))).)))))....... ( -23.70) >consensus GCAAUGAUAACCCAGCAAACAUGGCCCUGGCCCAAAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA_____ ...........(((((......(((....)))......(((........)))((((((((((....))))))..........((....)).....)))).)))))....... (-25.52 = -26.00 + 0.48)

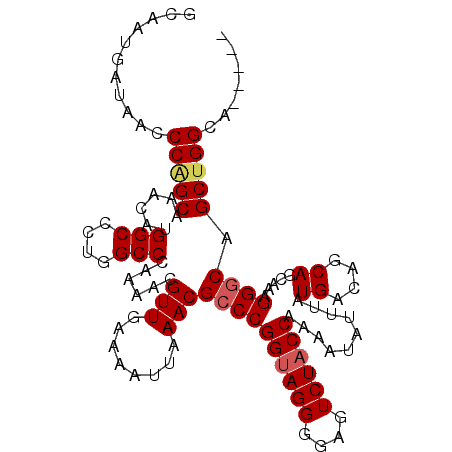

| Location | 13,727,403 – 13,727,512 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 94.14 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -25.22 |
| Energy contribution | -25.26 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13727403 109 + 23771897 AAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGGCAGCAGCAAAGGGCAGCUGGCA---------ACGGAGACAACGUUGCGUAUGCGUCAUAUUAAUGGA ...((((((.........((((((((((....))))))............((....))...)))).((..(((---------(((.......))))))....))......)))))).. ( -31.70) >DroSec_CAF1 134333 109 + 1 AAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA---------ACGGAGACAACGUUGCGUAUGCGUCAUAUUAAUGGA ...((((........)))).((((((((....))))))........((((((..((((........))))(((---------(((.......))))))......)))))).....)). ( -32.10) >DroSim_CAF1 145118 109 + 1 AAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA---------ACGGAGACAACGUUGCGUAUGCGUCAUAUUAAUGGA ...((((........)))).((((((((....))))))........((((((..((((........))))(((---------(((.......))))))......)))))).....)). ( -32.10) >DroEre_CAF1 141013 109 + 1 AAACGUUGAAAAUUAAACGGCCGGUAGGAGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA---------ACGGAGACAACGUUGCGUAUGCGUCAUAUUAAUGGA ...((((........)))).((((((((....))))))........((((((..((((........))))(((---------(((.......))))))......)))))).....)). ( -32.90) >DroYak_CAF1 144824 115 + 1 AAACGUUGAAAAUUAAACGCCCGGCAGGAGAGUCUACCAAAAUAUUUAUGACAGCAG---AGGGCAGCUGGCAAACUGGCAAACGGAGACAACGUUGCGUAUGCGUCAUAUUAAUGGA ...((((........)))).((((.(((....))).))........((((((.(((.---.(.(((((..((......))....(....)...))))).).))))))))).....)). ( -26.90) >consensus AAACGUUGAAAAUUAAACGCCCGGUAGGGGAGUCUACCAAAAUAUUUAUGACAGCAGCAAAGGGCAGCUGGCA_________ACGGAGACAACGUUGCGUAUGCGUCAUAUUAAUGGA ...((((........)))).((((((((....))))))........((((((.(((.....(.(((((.(.............)(....)...))))).).))))))))).....)). (-25.22 = -25.26 + 0.04)

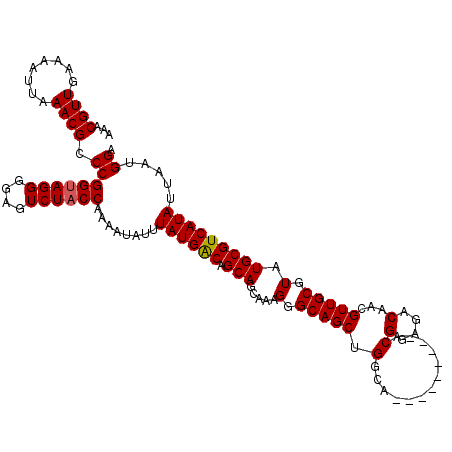

| Location | 13,727,403 – 13,727,512 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.14 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13727403 109 - 23771897 UCCAUUAAUAUGACGCAUACGCAACGUUGUCUCCGU---------UGCCAGCUGCCCUUUGCUGCUGCCAUAAAUAUUUUGGUAGACUCCCCUACCGGGCGUUUAAUUUUCAACGUUU ...........(((((....((((((.......)))---------)))((((.((.....)).))))...........(((((((......))))))))))))............... ( -26.90) >DroSec_CAF1 134333 109 - 1 UCCAUUAAUAUGACGCAUACGCAACGUUGUCUCCGU---------UGCCAGCUGCCCUUUGCUGCUGUCAUAAAUAUUUUGGUAGACUCCCCUACCGGGCGUUUAAUUUUCAACGUUU .((.....((((((......((((((.......)))---------)))((((........))))..))))))........(((((......)))))))(((((........))))).. ( -30.00) >DroSim_CAF1 145118 109 - 1 UCCAUUAAUAUGACGCAUACGCAACGUUGUCUCCGU---------UGCCAGCUGCCCUUUGCUGCUGUCAUAAAUAUUUUGGUAGACUCCCCUACCGGGCGUUUAAUUUUCAACGUUU .((.....((((((......((((((.......)))---------)))((((........))))..))))))........(((((......)))))))(((((........))))).. ( -30.00) >DroEre_CAF1 141013 109 - 1 UCCAUUAAUAUGACGCAUACGCAACGUUGUCUCCGU---------UGCCAGCUGCCCUUUGCUGCUGUCAUAAAUAUUUUGGUAGACUCUCCUACCGGCCGUUUAAUUUUCAACGUUU .((.....((((((......((((((.......)))---------)))((((........))))..))))))........(((((......))))))).((((........))))... ( -27.50) >DroYak_CAF1 144824 115 - 1 UCCAUUAAUAUGACGCAUACGCAACGUUGUCUCCGUUUGCCAGUUUGCCAGCUGCCCU---CUGCUGUCAUAAAUAUUUUGGUAGACUCUCCUGCCGGGCGUUUAAUUUUCAACGUUU .((.....(((((((((.((((((((.......)).))))..)).))).(((......---..)))))))))........(((((......)))))))(((((........))))).. ( -27.30) >consensus UCCAUUAAUAUGACGCAUACGCAACGUUGUCUCCGU_________UGCCAGCUGCCCUUUGCUGCUGUCAUAAAUAUUUUGGUAGACUCCCCUACCGGGCGUUUAAUUUUCAACGUUU ..............((....))(((((((................((.((((.((.....)).)))).))(((((...(((((((......)))))))..))))).....))))))). (-23.06 = -23.10 + 0.04)



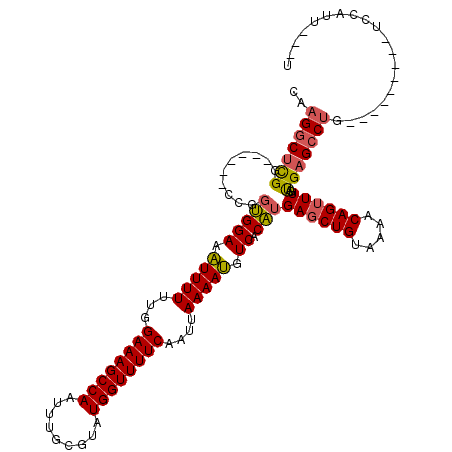
| Location | 13,727,512 – 13,727,617 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -26.71 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13727512 105 + 23771897 CAAGGCUCGG------CCGGUGGAAAUUUUUUGGAAAGCCAAUUUGCGUAUGGUUUUCAAUUAAAAUGUCACAUGAGCUGUAAAACAGUUUGAUGAGCCUG---------UCUCUUCCGU ..(((((((.------...((((..(((((...((((((((.........))))))))....))))).))))..((((((.....))))))..))))))).---------.......... ( -29.60) >DroPse_CAF1 127527 117 + 1 CAAGGCUCGGGCUCGGGCUGUGGACAUUUUUUGGAAAGCCAAUUUGCGUAUGGUUUUCAAUUAAAAUGUCACAUGAGCUGUAAAACAGUUUGAUGAGCCUGUCGUCCAGGGCCAGG---U ...(((((((((.(((((((((((((((((...((((((((.........))))))))....)))))))).)))((((((.....))))))....))))))..)))).)))))...---. ( -48.90) >DroEre_CAF1 141122 105 + 1 CAAGGCUCGG------CCGGUGGAAAUUUUUUGGAAAGCCAAUUUGCGUAUGGUUUUCAAUUAAAAUGUCACAUGAGCUGUAAAACAGUUUGAUGAGCCUG---------UCCCCUUCGU ..(((((((.------...((((..(((((...((((((((.........))))))))....))))).))))..((((((.....))))))..))))))).---------.......... ( -29.60) >DroYak_CAF1 144939 105 + 1 CAAGGCUCGG------GCGGUGGAAAUUUUUUGGAAAGCCAAUUUGCGUAUGGUUUUCAAUUAAAAUGUCACAUGAGCUGUAAAACAGUUUGAUGAGCCUG---------UCUCUUCCGU ..(((((((.------...((((..(((((...((((((((.........))))))))....))))).))))..((((((.....))))))..))))))).---------.......... ( -29.60) >DroAna_CAF1 195338 111 + 1 CGUGGCUUGG------CCACAGGAUGUUUUUUGGAAAGCCAAUUUGAGUAUGGUUUUCAAUUAAAACGUCACUCGAGCUGUAAAACAGGUUGACGUGGCUACAUUCGAGGUCCAUU---U .(((((...)------)))).(((((((((...((((((((.........))))))))....)))))(((((((((.(((.....))).)))).)))))..........))))...---. ( -38.60) >DroPer_CAF1 128909 117 + 1 CAAGGCUCGGGCUCGGGCUGUGGACAUUUUUUGGAAAGCCAAUUUGCGUAUGGUUUUCAAUUAAAAUGUCACAUGAGCUGUAAAACAGUUUGAUGAGCCUGUCGUCCAGGGCCAGG---U ...(((((((((.(((((((((((((((((...((((((((.........))))))))....)))))))).)))((((((.....))))))....))))))..)))).)))))...---. ( -48.90) >consensus CAAGGCUCGG______CCGGUGGAAAUUUUUUGGAAAGCCAAUUUGCGUAUGGUUUUCAAUUAAAAUGUCACAUGAGCUGUAAAACAGUUUGAUGAGCCUG_________UCCAUU___U ..(((((((..........(((((.(((((...((((((((.........))))))))....))))).)).)))((((((.....))))))..))))))).................... (-26.71 = -26.72 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:15 2006