
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,725,031 – 13,725,121 |
| Length | 90 |
| Max. P | 0.939547 |

| Location | 13,725,031 – 13,725,121 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -18.71 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13725031 90 + 23771897 GCAACGUCCCAC--GCUUUUGGAGUGCGAAUUUGCGUAAAUAUAAUUUAUUUACGCAAUUGCAAAGGA----------CCCGAGAAGGUUUCG---------------CAGGAUUAU .....((((...--((........(((((..(((((((((((.....))))))))))))))))..(((----------.((.....)).))))---------------).))))... ( -25.00) >DroPse_CAF1 124684 100 + 1 GCCAUGUCCCACGCGCUUUUGGAGUGCGAAUUUGCGUAAAUAUAAUUUAUUUACGCAAUGGCAAAGGA----------UCCGAGAAGGUUUCG--GCAUAGA-G----CAGGCACAU (((..((((..(((((((...)))))))...(((((((((((.....))))))))))).......)))----------)((((((...)))))--)......-.----..))).... ( -34.10) >DroEre_CAF1 138619 90 + 1 GCAACGUCCCAC--GCUUUUGGAGUGCGAAUUUGCGUAAAUAUAAUUUAUUUACGCAAUUGCAAGGGA----------CCGGAGAAGGCUUCG---------------CAGGAUUAU ((...(((((..--((((...))))((((..(((((((((((.....)))))))))))))))..))))----------).((((....)))))---------------)........ ( -30.90) >DroYak_CAF1 142366 90 + 1 GCAACGUCCCAC--GCUUUUGGAGUGCGAAUUUGCGUAAAUAUAAUUUAUUUACGCAAUUGCCAAGGA----------CCGGAGAAGGCUUCG---------------CAGGAUUAU ((...((((...--((((...))))((((..(((((((((((.....)))))))))))))))...)))----------).((((....)))))---------------)........ ( -25.80) >DroAna_CAF1 192847 115 + 1 GCGAGGCCCCCC--GUUUUUGGAGUGCGAAUUUGCGUAAAUAUAAUUUAUUUACGCAAUGGCCAACGAGCCAUGGCAGCCCUGGAAGGCUCACCCGGACGGAGGCAGUCAUGAUUAU ....((((..((--(....))).........(((((((((((.....))))))))))).)))).......((((((.((((((...((.....))...))).))).))))))..... ( -39.90) >DroPer_CAF1 126076 100 + 1 GCCAUGUCCCACGCGCUUUUGGAGUGCGAAUUUGCGUAAAUAUAAUUUAUUUACGCAAUGGCAAAGGA----------UCCGAGAAGGUUUCG--GCAUAGA-G----CAGGCACAU (((..((((..(((((((...)))))))...(((((((((((.....))))))))))).......)))----------)((((((...)))))--)......-.----..))).... ( -34.10) >consensus GCAACGUCCCAC__GCUUUUGGAGUGCGAAUUUGCGUAAAUAUAAUUUAUUUACGCAAUGGCAAAGGA__________CCCGAGAAGGCUUCG_______________CAGGAUUAU (((((((((((........))).).)))...(((((((((((.....)))))))))))))))...................(((.....)))......................... (-18.71 = -18.02 + -0.69)


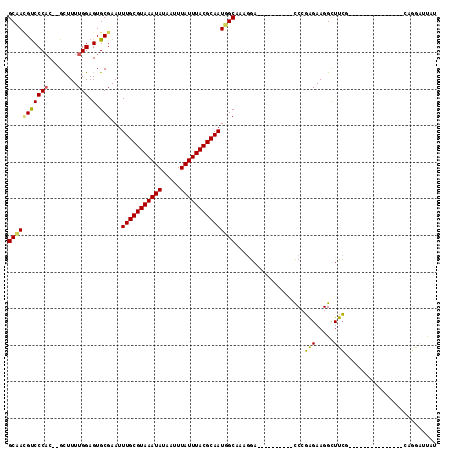
| Location | 13,725,031 – 13,725,121 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939547 |
| Prediction | RNA |
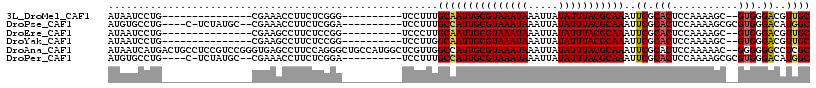
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13725031 90 - 23771897 AUAAUCCUG---------------CGAAACCUUCUCGGG----------UCCUUUGCAAUUGCGUAAAUAAAUUAUAUUUACGCAAAUUCGCACUCCAAAAGC--GUGGGACGUUGC ....(((..---------------((..((((....)))----------)....((((((((((((((((.....))))))))).)))).))).........)--)..)))...... ( -24.90) >DroPse_CAF1 124684 100 - 1 AUGUGCCUG----C-UCUAUGC--CGAAACCUUCUCGGA----------UCCUUUGCCAUUGCGUAAAUAAAUUAUAUUUACGCAAAUUCGCACUCCAAAAGCGCGUGGGACAUGGC ....(((((----.-.((((((--(((.......))(((----------.....(((..(((((((((((.....)))))))))))....))).)))....).))))))..)).))) ( -29.00) >DroEre_CAF1 138619 90 - 1 AUAAUCCUG---------------CGAAGCCUUCUCCGG----------UCCCUUGCAAUUGCGUAAAUAAAUUAUAUUUACGCAAAUUCGCACUCCAAAAGC--GUGGGACGUUGC ....(((..---------------((..(((......))----------)....((((((((((((((((.....))))))))).)))).))).........)--)..)))...... ( -24.60) >DroYak_CAF1 142366 90 - 1 AUAAUCCUG---------------CGAAGCCUUCUCCGG----------UCCUUGGCAAUUGCGUAAAUAAAUUAUAUUUACGCAAAUUCGCACUCCAAAAGC--GUGGGACGUUGC ....(((..---------------(((((((........----------.....)))..(((((((((((.....))))))))))).)))((.........))--)..)))...... ( -25.22) >DroAna_CAF1 192847 115 - 1 AUAAUCAUGACUGCCUCCGUCCGGGUGAGCCUUCCAGGGCUGCCAUGGCUCGUUGGCCAUUGCGUAAAUAAAUUAUAUUUACGCAAAUUCGCACUCCAAAAAC--GGGGGGCCUCGC .............(((((((...((.((((((....)))))((..(((((....)))))(((((((((((.....)))))))))))....)).).))....))--)))))....... ( -40.20) >DroPer_CAF1 126076 100 - 1 AUGUGCCUG----C-UCUAUGC--CGAAACCUUCUCGGA----------UCCUUUGCCAUUGCGUAAAUAAAUUAUAUUUACGCAAAUUCGCACUCCAAAAGCGCGUGGGACAUGGC ....(((((----.-.((((((--(((.......))(((----------.....(((..(((((((((((.....)))))))))))....))).)))....).))))))..)).))) ( -29.00) >consensus AUAAUCCUG_______________CGAAACCUUCUCGGG__________UCCUUUGCAAUUGCGUAAAUAAAUUAUAUUUACGCAAAUUCGCACUCCAAAAGC__GUGGGACGUUGC .......................................................(((((((((((((((.....)))))))))))..((.(((...........))).))..)))) (-16.56 = -16.42 + -0.14)
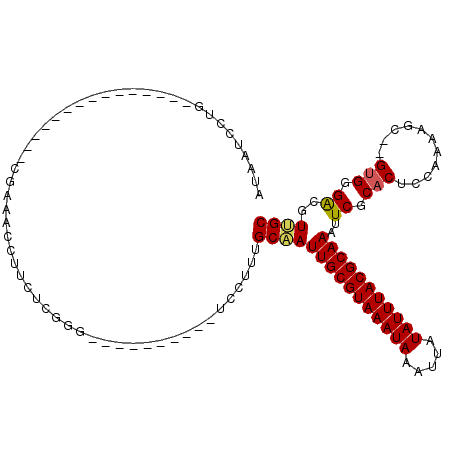

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:11 2006