| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,723,474 – 13,723,564 |
| Length | 90 |
| Max. P | 0.799264 |

| Location | 13,723,474 – 13,723,564 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -15.01 |
| Energy contribution | -16.45 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
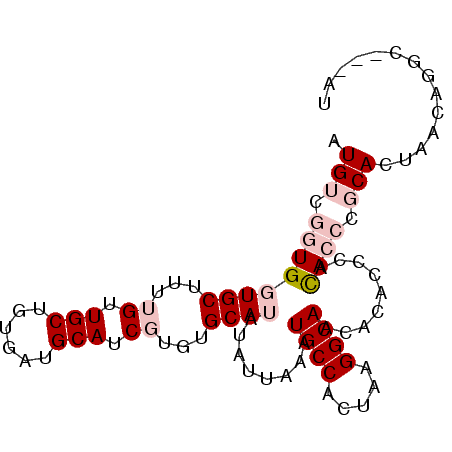
>3L_DroMel_CAF1 13723474 90 + 23771897 AUGUCGGUGGUGCUUUUGUUGCUGUGAUGCAUCGUGUGCAUAUAUUAAAUGCCACUAAGGCAACACACCCACACCCGCACUAACAGGC---AC .((((..((((((...(((.(.((((.(((...(((.((((.......)))))))....))).)))).).)))...))))))...)))---). ( -30.00) >DroPse_CAF1 122401 86 + 1 UUG---CUGCUGCUACCGCUGC----AUGCAUCAUGUGCAUAUAUUAAAUGCCACUAAGGCAACACUUGCAUACGCACACGCACAGCCUCUAU .((---((((.((....)).))----).)))...(((((..........((((.....)))).....(((....)))...)))))........ ( -23.10) >DroSec_CAF1 130431 90 + 1 AUGUCGGUGGUGCUUUUGUUGCUGUGAUGCAUCGUGUGCAUAUAUUAAAUGCCACUAAGGCAACACACCCACACCCGCACUAACAGGC---AC .((((..((((((...(((.(.((((.(((...(((.((((.......)))))))....))).)))).).)))...))))))...)))---). ( -30.00) >DroSim_CAF1 141219 90 + 1 AUGUCGGUGGUGCUUUUGUUGCUGUGAUGCAUCGUGUGCAUAUAUUAAAUGCCACUAAGGCAACACACCCACACCCGCACUAACAGGC---AA .((((..((((((...(((.(.((((.(((...(((.((((.......)))))))....))).)))).).)))...))))))...)))---). ( -30.30) >DroEre_CAF1 137070 88 + 1 AUGUCGGUGGUGCUUUUGUUGCUGUGAUGCAUCGUGUGCAUAUAUUAAAUGCCACUAAGGCAACACACA--CACCCGCACUAACAGGC---AU (((((..((((((...(((.(.((((.(((...(((.((((.......)))))))....))).))))))--))...))))))...)))---)) ( -29.20) >DroPer_CAF1 123766 86 + 1 UUG---CUGCUGCUACCGCUGC----AUGCAUCAUGUGCAUAUAUUAAAUGCCACUAAGGCAACACUUGCAUACGCACACGCACAGCCUCUAU .((---((((.((....)).))----).)))...(((((..........((((.....)))).....(((....)))...)))))........ ( -23.10) >consensus AUGUCGGUGGUGCUUUUGUUGCUGUGAUGCAUCGUGUGCAUAUAUUAAAUGCCACUAAGGCAACACACCCACACCCGCACUAACAGGC___AU .(((.((((((((...((.(((......))).))...))))........((((.....)))).........)))).))).............. (-15.01 = -16.45 + 1.44)

| Location | 13,723,474 – 13,723,564 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.53 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
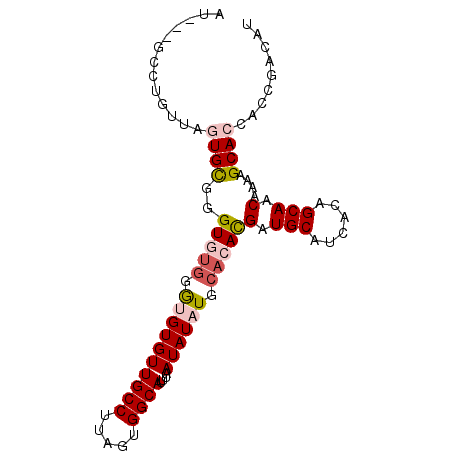
>3L_DroMel_CAF1 13723474 90 - 23771897 GU---GCCUGUUAGUGCGGGUGUGGGUGUGUUGCCUUAGUGGCAUUUAAUAUAUGCACACGAUGCAUCACAGCAACAAAAGCACCACCGACAU ..---((((((....))))))((.((((.((.((....(((((((.......)))).)))(.(((......))).)....)))))))).)).. ( -29.40) >DroPse_CAF1 122401 86 - 1 AUAGAGGCUGUGCGUGUGCGUAUGCAAGUGUUGCCUUAGUGGCAUUUAAUAUAUGCACAUGAUGCAU----GCAGCGGUAGCAGCAG---CAA ......(((((.((((((((((((.((((((..(....)..))))))..)))))))))))).(((.(----((....))))))))))---).. ( -34.50) >DroSec_CAF1 130431 90 - 1 GU---GCCUGUUAGUGCGGGUGUGGGUGUGUUGCCUUAGUGGCAUUUAAUAUAUGCACACGAUGCAUCACAGCAACAAAAGCACCACCGACAU ..---((((((....))))))((.((((.((.((....(((((((.......)))).)))(.(((......))).)....)))))))).)).. ( -29.40) >DroSim_CAF1 141219 90 - 1 UU---GCCUGUUAGUGCGGGUGUGGGUGUGUUGCCUUAGUGGCAUUUAAUAUAUGCACACGAUGCAUCACAGCAACAAAAGCACCACCGACAU ..---((((((....))))))((.((((.((.((....(((((((.......)))).)))(.(((......))).)....)))))))).)).. ( -29.40) >DroEre_CAF1 137070 88 - 1 AU---GCCUGUUAGUGCGGGUG--UGUGUGUUGCCUUAGUGGCAUUUAAUAUAUGCACACGAUGCAUCACAGCAACAAAAGCACCACCGACAU ..---((((((....))))))(--(((.((((((....(((((((.......)))).)))((....))...))))))...))))......... ( -26.50) >DroPer_CAF1 123766 86 - 1 AUAGAGGCUGUGCGUGUGCGUAUGCAAGUGUUGCCUUAGUGGCAUUUAAUAUAUGCACAUGAUGCAU----GCAGCGGUAGCAGCAG---CAA ......(((((.((((((((((((.((((((..(....)..))))))..)))))))))))).(((.(----((....))))))))))---).. ( -34.50) >consensus AU___GCCUGUUAGUGCGGGUGUGGGUGUGUUGCCUUAGUGGCAUUUAAUAUAUGCACACGAUGCAUCACAGCAACAAAAGCACCACCGACAU .............((((..(((((.((((((((((.....))))....)))))).)))))(.(((......))).)....))))......... (-18.86 = -19.53 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:09 2006