
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,704,963 – 13,705,099 |
| Length | 136 |
| Max. P | 0.775669 |

| Location | 13,704,963 – 13,705,060 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 62.57 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -14.75 |
| Energy contribution | -14.72 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13704963 97 - 23771897 UGCACGGAAUGCCAUGGCGAACUUGCCAA----ACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAUUGUCUGGCUGGCUGGCUACAU------- .((.(((...((((.(((((.((((((..----.(((.(((((((......)))))))....))).....))))))...)))))))))...))))).....------- ( -39.20) >DroSec_CAF1 112185 101 - 1 UGCACGGAAUGCCAUGGCAAACUUGCCAAACUUACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAUUGUCUGGCUGGCUGGCUACAU------- .((.(((...((((.(((((.((((((.((.((.(((.(((((((......)))))))....))))))).))))))...)))))))))...))))).....------- ( -39.40) >DroSim_CAF1 122990 100 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNUGCCAGGAGCAGGGAAUUCGGCAAGGGA-UGUCUGGCUGGCUGGCUACAU------- ....................................................(((((.(((.((..((((......)))-)..)).)))..))))).....------- ( -15.60) >DroYak_CAF1 121980 97 - 1 UGCACGGAAUGCCAUGGCAAACUUGCCAA----ACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAUUGUCUGGCUGGCUGGCUACAU------- .((.(((...((((.(((((.((((((..----.(((.(((((((......)))))))....))).....))))))...)))))))))...))))).....------- ( -39.50) >DroAna_CAF1 169358 99 - 1 UGGAAAGAAACCUCUGCCAAACUUGCCAG----ACCCACUUGGUAUGUCUCUGCCGGUUAGAGCGAGUUAU-----GGGUUAGAUGGUUAGACGUCUCUAUGCGAGGG ..........((((..(((((((((((.(----(((.....((((......)))))))).).))))))).)-----))((.(((((......)))))....)))))). ( -29.20) >consensus UGCACGGAAUGCCAUGGCAAACUUGCCAA____ACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAUUGUCUGGCUGGCUGGCUACAU_______ (((..((....))...))).((.((((((..........)))))).))....(((((.((((((.(((((......))))).))).)))..)))))............ (-14.75 = -14.72 + -0.03)

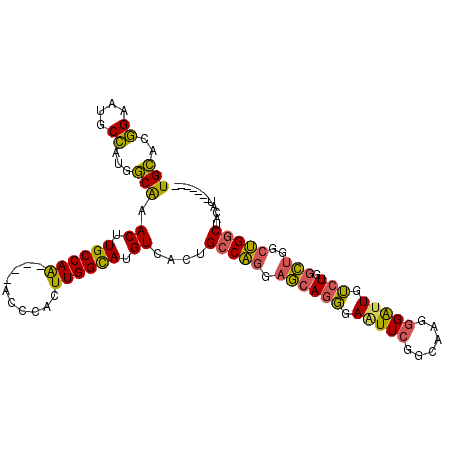

| Location | 13,704,984 – 13,705,099 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -21.87 |
| Energy contribution | -24.40 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13704984 115 + 23771897 AUCCCUUGCCGAAUUCCCUGCUCCUGGCAGUGACAUGCCAAGUGGGU----UUGGCAAGUUCGCCAUGGCAUUCCGUGCACUUCCCGCCACCCGCACACGCACUUGGCAUGUAAAAUAG ...(.((((((.............)))))).)(((((((((((((((----.(((.((((..((.((((....)))))))))).)))..))))((....)))))))))))))....... ( -39.12) >DroPse_CAF1 102846 89 + 1 -----UUGC------CUCUGCUGCUGGCAGUGACACGACAAGUGGGU----UUGGCAACUUUGCCAUAGCAUACUCUUCAG----CG----CCUCACAU-------GCAUGCAAAAUAG -----((((------...(((((..(((..((((((.....))).((----((((((....))))).))).......))).----.)----))...)).-------))).))))..... ( -22.90) >DroSec_CAF1 112206 119 + 1 AUCCCUUGCCGAAUUCCCUGCUCCUGGCAGUGACAUGCCAAGUGGGUAAGUUUGGCAAGUUUGCCAUGGCAUUCCGUGCACUUCCCGCCACCCGCACACGCACUUGGCAUGCAAAAUAG ....(((((((((((.((..((..(((((......)))))))..))..)))))))))))((((((((((....)))))........((((...((....))...))))..))))).... ( -42.80) >DroEre_CAF1 118445 115 + 1 AUCCCUUGCCGAAUUCCCUGCUCCUGGCAGUGACAUGCCAAGUGGGU----UUGGCAAGUUUGCCAUGGCAUUCCGUGCACUUCCCGCCACCCGCACACGCACUUGGCAUGUAAAAUAG ...(.((((((.............)))))).)(((((((((((((((----.(((.((((..((.((((....)))))))))).)))..))))((....)))))))))))))....... ( -39.12) >DroYak_CAF1 122001 115 + 1 AUCCCUUGCCGAAUUCCCUGCUCCUGGCAGUGACAUGCCAAGUGGGU----UUGGCAAGUUUGCCAUGGCAUUCCGUGCACUUCCCAUCACCCGCACACGCACUUGGCAUGUAAAAUAG ...(.((((((.............)))))).)(((((((((((((((----.(((.((((..((.((((....)))))))))).)))..))))((....)))))))))))))....... ( -40.32) >DroPer_CAF1 104264 89 + 1 -----UUGC------CUCUGCUGCUGGCAGUGACACGACAAGUGGGU----UUGGCAACUUUGCCAUAGCAUACUCUUCAG----CG----CCUCACAU-------GCAUGCAAAAUAG -----((((------...(((((..(((..((((((.....))).((----((((((....))))).))).......))).----.)----))...)).-------))).))))..... ( -22.90) >consensus AUCCCUUGCCGAAUUCCCUGCUCCUGGCAGUGACAUGCCAAGUGGGU____UUGGCAAGUUUGCCAUGGCAUUCCGUGCACUUCCCGCCACCCGCACACGCACUUGGCAUGCAAAAUAG .................((((.....))))((.(((((((((((.((.....((((......))))..(((.....)))..................)).)))))))))))))...... (-21.87 = -24.40 + 2.53)

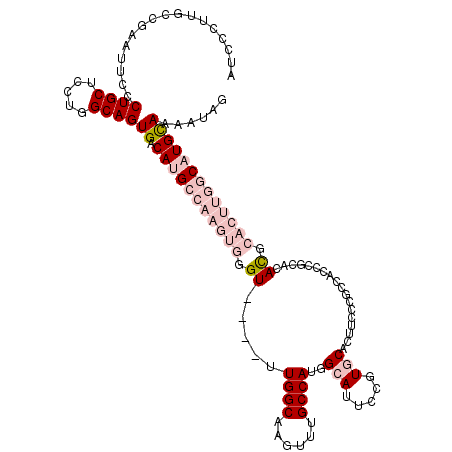

| Location | 13,704,984 – 13,705,099 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -24.61 |
| Energy contribution | -27.67 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13704984 115 - 23771897 CUAUUUUACAUGCCAAGUGCGUGUGCGGGUGGCGGGAAGUGCACGGAAUGCCAUGGCGAACUUGCCAA----ACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAU .......(((((((((((((....))((((((((((...(((..((....))...)))..))))))..----)))))))))))))))(..((((.(((........)))))))..)... ( -42.90) >DroPse_CAF1 102846 89 - 1 CUAUUUUGCAUGC-------AUGUGAGG----CG----CUGAAGAGUAUGCUAUGGCAAAGUUGCCAA----ACCCACUUGUCGUGUCACUGCCAGCAGCAGAG------GCAA----- .....((((.(((-------.(((..((----((----.(((...........(((((....))))).----...(((.....)))))).)))).))))))...------))))----- ( -25.50) >DroSec_CAF1 112206 119 - 1 CUAUUUUGCAUGCCAAGUGCGUGUGCGGGUGGCGGGAAGUGCACGGAAUGCCAUGGCAAACUUGCCAAACUUACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAU ...((((((((((((((((.(.(((.(..(((((((...(((..((....))...)))..)))))))..).)))))))))))))).((.((((.....)))).)).....))))))... ( -46.90) >DroEre_CAF1 118445 115 - 1 CUAUUUUACAUGCCAAGUGCGUGUGCGGGUGGCGGGAAGUGCACGGAAUGCCAUGGCAAACUUGCCAA----ACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAU .......(((((((((((((....))((((((((((...(((..((....))...)))..))))))..----)))))))))))))))(..((((.(((........)))))))..)... ( -43.60) >DroYak_CAF1 122001 115 - 1 CUAUUUUACAUGCCAAGUGCGUGUGCGGGUGAUGGGAAGUGCACGGAAUGCCAUGGCAAACUUGCCAA----ACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAU .......(((((((((((((....))((((..(((.((((((..((....))...))..)))).))).----)))))))))))))))(..((((.(((........)))))))..)... ( -41.80) >DroPer_CAF1 104264 89 - 1 CUAUUUUGCAUGC-------AUGUGAGG----CG----CUGAAGAGUAUGCUAUGGCAAAGUUGCCAA----ACCCACUUGUCGUGUCACUGCCAGCAGCAGAG------GCAA----- .....((((.(((-------.(((..((----((----.(((...........(((((....))))).----...(((.....)))))).)))).))))))...------))))----- ( -25.50) >consensus CUAUUUUACAUGCCAAGUGCGUGUGCGGGUGGCGGGAAGUGCACGGAAUGCCAUGGCAAACUUGCCAA____ACCCACUUGGCAUGUCACUGCCAGGAGCAGGGAAUUCGGCAAGGGAU .......(((((((((((((((((((............)))))))........(((((....)))))........))))))))))))..((((.....))))................. (-24.61 = -27.67 + 3.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:48 2006