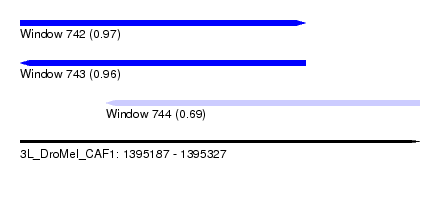
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,395,187 – 1,395,327 |
| Length | 140 |
| Max. P | 0.974453 |

| Location | 1,395,187 – 1,395,287 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -17.96 |
| Energy contribution | -17.88 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1395187 100 + 23771897 ACAGGAUCGCCUGAAUCACGAGCAUAAGA--------ACACGGAGCAUCAUCGUCCAGAUUUGCAAUCCAAGCCGAAUGUGAUCAUCCUGGGCAUUGAUUCCACAUCC .((((....))))................--------....(((..((((..((((((..(..((.((......)).))..).....))))))..)))))))...... ( -23.00) >DroSec_CAF1 50126 100 + 1 ACAGGAUCGCCUGAAUCACGGGCAUAAGA--------GUCCGGAGGAUCAUCGGCCAGAUUUGGAGUCCAAGCCGAAUGUGAUCAUCCUGGGCAUUGAUUCCAUAUCC ...((((((((((.....)))))......--------(((((((.((((((((((..(((.....)))...)))))...))))).))).))))...)))))....... ( -36.30) >DroSim_CAF1 47780 100 + 1 ACAGGAUCGCCUGAAUCACGGGCAUAAGA--------GUCCGGAGGAUCAUCGUCCAGAUUUGGAGUCCAAGCCCAAUGUGAUCAUCCUGGGCAUUGAUUCCACAUCC .((((....))))..((.(((((......--------)))))))((((....)))).(((.(((((((...((((((((....)))..)))))...))))))).))). ( -37.00) >DroEre_CAF1 51835 88 + 1 GCAGGAUCGCCUGAAGAA---A-----------------CCAGAGGAUCCUCCCCCAGAUCUGGAGUCCAAGCCGAAUGUGAUCAUCCUGGGCAUAGACUCCACAUCC ..((((((..(((.....---.-----------------.)))..))))))......(((.(((((((...(((.((((....)))..).)))...))))))).))). ( -28.50) >DroYak_CAF1 53462 91 + 1 GCAGAAUCGCCUGAAGCACAGG-----------------CAGAAGGAUCCUCCUUCAGAUGUGGAGUCCAAGCCGAAUGUGAUCAUCCUGGGCAUUGACUCCACAUCG ........(((((.....))))-----------------).(((((.....))))).(((((((((((...(((.((((....)))..).)))...))))))))))). ( -40.70) >DroPer_CAF1 77720 106 + 1 CCAGGAUCAUUUGAGCUACGCGGACCUGGUGCGCGCCUUCCGCCUGCUCCACGAC--GAGGAGGAGCCCAAGCCGCAUGUCAUCAUCCUGGGCAUUGACUCCACCUCG ............((((...(((((...(((....))).)))))..)))).....(--((((.((((..((((((.((.(.......).))))).))).)))).))))) ( -37.80) >consensus ACAGGAUCGCCUGAAUCACGGGCAUAAGA_________UCCGGAGGAUCAUCGUCCAGAUUUGGAGUCCAAGCCGAAUGUGAUCAUCCUGGGCAUUGACUCCACAUCC .((((....))))............................................(((.(((((((...(((.((((....)))..).)))...))))))).))). (-17.96 = -17.88 + -0.08)

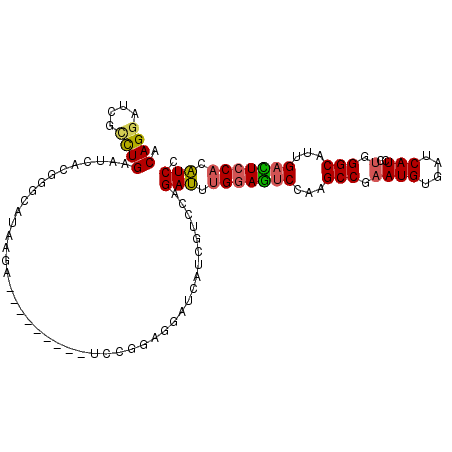
| Location | 1,395,187 – 1,395,287 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -20.19 |
| Energy contribution | -20.83 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1395187 100 - 23771897 GGAUGUGGAAUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGAUUGCAAAUCUGGACGAUGAUGCUCCGUGU--------UCUUAUGCUCGUGAUUCAGGCGAUCCUGU ((((((((.(((.........)))..)))))))).....(((((((....(((((..((((.((.......--------......))))))..))))))))))))... ( -27.22) >DroSec_CAF1 50126 100 - 1 GGAUAUGGAAUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCAAAUCUGGCCGAUGAUCCUCCGGAC--------UCUUAUGCCCGUGAUUCAGGCGAUCCUGU ((((...((((((.......(((.(((((...(((((((((...))).....))))))))))).)))((.(--------......).)).)))))).....))))... ( -31.10) >DroSim_CAF1 47780 100 - 1 GGAUGUGGAAUCAAUGCCCAGGAUGAUCACAUUGGGCUUGGACUCCAAAUCUGGACGAUGAUCCUCCGGAC--------UCUUAUGCCCGUGAUUCAGGCGAUCCUGU ((((.((((.((...((((((..........))))))...)).))))..((((((.(.....).)))))).--------.....((((.(.....).))))))))... ( -31.00) >DroEre_CAF1 51835 88 - 1 GGAUGUGGAGUCUAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCAGAUCUGGGGGAGGAUCCUCUGG-----------------U---UUCUUCAGGCGAUCCUGC ((((.(((((((((.(((..(((((....)))))))).))))))))).))))...(.((((((.(((((-----------------.---....))))).)))))).) ( -42.00) >DroYak_CAF1 53462 91 - 1 CGAUGUGGAGUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCACAUCUGAAGGAGGAUCCUUCUG-----------------CCUGUGCUUCAGGCGAUUCUGC .(((((((((((...(((..(((((....))))))))...))))))))))).(((((.....)))))((-----------------((((.....))))))....... ( -44.00) >DroPer_CAF1 77720 106 - 1 CGAGGUGGAGUCAAUGCCCAGGAUGAUGACAUGCGGCUUGGGCUCCUCCUC--GUCGUGGAGCAGGCGGAAGGCGCGCACCAGGUCCGCGUAGCUCAAAUGAUCCUGG (((((.((((((...(((..(.(((....))).))))...)))))).))))--)((((.(((((.(((((.((......))...))))).).))))..))))...... ( -44.70) >consensus GGAUGUGGAAUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCAAAUCUGGACGAGGAUCCUCCGGA_________UCUUAUGCCCGUGAUUCAGGCGAUCCUGU .(((.(((((((...(((..(((((....))))))))...))))))).)))............................................((((....)))). (-20.19 = -20.83 + 0.64)

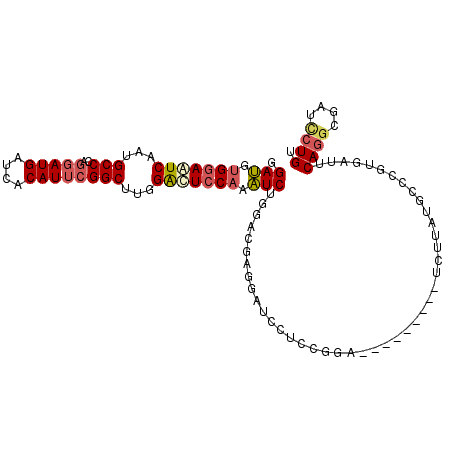
| Location | 1,395,217 – 1,395,327 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -29.83 |
| Energy contribution | -31.67 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
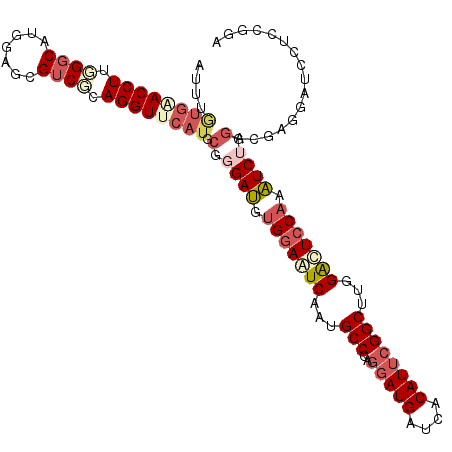
>3L_DroMel_CAF1 1395217 110 - 23771897 AUUUGUGAACCUUGGGCAUGGAGCGUCUCAGGUUCAUGCGGGAUGUGGAAUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGAUUGCAAAUCUGGACGAUGAUGCUCCGUG ................(((((((((((((.((((((.((.((((((((.(((.........)))..)))))))).)).)))))).((....))...)).))))))))))) ( -36.90) >DroSec_CAF1 50156 110 - 1 AUUUGUGAACCUUGGGCAUGGAGCGUCGCAGGUUCAUGCGGGAUAUGGAAUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCAAAUCUGGCCGAUGAUCCUCCGGA .........((.(((((((.((...((.((.((((.....)))).)))).)).)))))))(((.(((((...(((((((((...))).....))))))))))).))))). ( -37.90) >DroSim_CAF1 47810 110 - 1 AUUUGUGAACCUUGGGCAUGGAGCGUCGCAGGUUCAUGCGGGAUGUGGAAUCAAUGCCCAGGAUGAUCACAUUGGGCUUGGACUCCAAAUCUGGACGAUGAUCCUCCGGA ....(((((((((((((.....)).))).)))))))).((((((.((((.((...((((((..........))))))...)).)))).))).(((......))).))).. ( -36.80) >DroEre_CAF1 51854 109 - 1 AUUUAUGCACCUUGGGCAUAGAGCGUCUCAGGUUCAUGCGGGAUGUGGAGUCUAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCAGAUCUGGGGGAGGAUCCUCUGG- ...((((.((((.((((.......)))).)))).))))(.((((.(((((((((.(((..(((((....)))))))).))))))))).)))).).((((....))))..- ( -45.80) >DroYak_CAF1 53484 109 - 1 AUUUAUGCACCUUGGGCAUAGAGCGUCUUAGGUUCAUGCGCGAUGUGGAGUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCACAUCUGAAGGAGGAUCCUUCUG- ...((((.((((.((((.......)))).)))).))))...(((((((((((...(((..(((((....))))))))...))))))))))).(((((.....)))))..- ( -46.30) >DroPer_CAF1 77758 108 - 1 AGCGCAGGACCUGCGGCAUGGUGCGACGCAGGUUGAUGCGCGAGGUGGAGUCAAUGCCCAGGAUGAUGACAUGCGGCUUGGGCUCCUCCUC--GUCGUGGAGCAGGCGGA .(((((.((((((((((.....))..))))))))..)))))((((.((((((...(((..(.(((....))).))))...)))))).))))--(((........)))... ( -52.40) >consensus AUUUGUGAACCUUGGGCAUGGAGCGUCGCAGGUUCAUGCGGGAUGUGGAAUCAAUGCCCAGGAUGAUCACAUUCGGCUUGGACUCCAAAUCUGGACGAGGAUCCUCCGGA ....((((((((.((((.......)))).)))))))).(.((((.(((((((...(((..(((((....))))))))...))))))).)))).)................ (-29.83 = -31.67 + 1.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:31 2006