
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,704,177 – 13,704,292 |
| Length | 115 |
| Max. P | 0.733361 |

| Location | 13,704,177 – 13,704,292 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.46 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13704177 115 - 23771897 CAAAUGAAAUCACACUGACCAGGCCCUAGAAUCAUUUUUCAAAAAUCAUUCACAGCCAAGUGUGCAUUCAAUCGUGCAGCACUCUC----UGCCUCCAAGCCCCAAGCCCGGC-CGAAAC ....((((..((((((.....(((....((((.((((.....)))).))))...))).))))))..)))).(((.((((......)----)))......(((........)))-)))... ( -23.10) >DroPse_CAF1 101896 112 - 1 CGAAUGAAAUCACACUGACCAGGCUGUAGAAUCAUUUUUCAAAAAUCAUUCACAGCCAAGUGUGCAUUCAAUCGUGUACCACUCUCCCUAUUUCUCUCU-------ACCCCUU-CUACCC (((.((((..((((((.....((((((.((((.((((.....)))).)))))))))).))))))..)))).))).........................-------.......-...... ( -25.40) >DroEre_CAF1 117627 109 - 1 CAAAUGAAAUCACACUGACCAGGCCCUAGAAUCAUUUUUCAAAAAUCAUUCACAGCCAAGUGUGCAUUCAAUCGUGCAGCACUCUC----UGCCCCCGA-------ACCCGGCCCAAAAC ....((((..((((((.....(((....((((.((((.....)))).))))...))).))))))..)))).....((((......)----)))..(((.-------...)))........ ( -20.70) >DroYak_CAF1 121128 108 - 1 CAAAUGAAAUCACACUGACCAGGCCCUAGAAUCAUUUUUCAAAAAUCAUUCACAGCCAAGUGUGCAUUCAAUCGUGCUGCACUCUC----UGCCUCCAA-------GCCCGUC-CGAAAU ..............(.(((..(((....((((.((((.....)))).)))).(((...(((((((((......)))).)))))..)----)).......-------))).)))-.).... ( -19.10) >DroAna_CAF1 167844 107 - 1 CAAAUGAAAUCACACUGACCAAGCCCUAGAAUCAUUUUUCAAAAAUCAUUCACAGCCAAGUGUGCAUUCAAUCGUGCACUACACUCC---CGCCUCCGC-------CUCCGCC-U--CCC ....((((..((((((......((....((((.((((.....)))).))))...))..))))))..))))...(((.....)))...---.........-------.......-.--... ( -12.60) >DroPer_CAF1 103314 112 - 1 CGAAUGAAAUCACACUGACCAGGCUGUAGAAUCAUUUUUCAAAAAUCAUUCACAGCCAAGUGUGCAUUCAAUCGUGUACCACUCUCCCUCUUUCUCUCU-------ACCCCUU-CUACCC (((.((((..((((((.....((((((.((((.((((.....)))).)))))))))).))))))..)))).))).........................-------.......-...... ( -25.40) >consensus CAAAUGAAAUCACACUGACCAGGCCCUAGAAUCAUUUUUCAAAAAUCAUUCACAGCCAAGUGUGCAUUCAAUCGUGCACCACUCUC____UGCCUCCAA_______ACCCGUC_CGAAAC ....((((..((((((.....(((....((((.((((.....)))).))))...))).))))))..))))...(((...)))...................................... (-14.09 = -14.12 + 0.03)

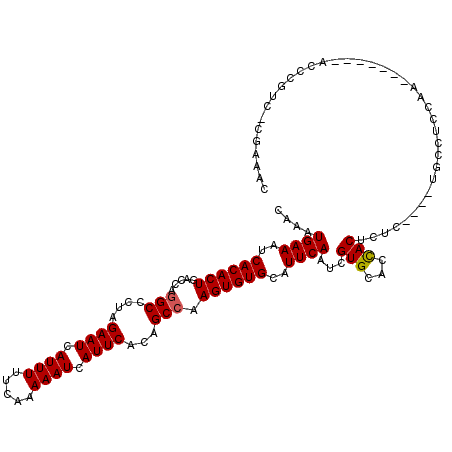
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:44 2006