
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,702,861 – 13,702,954 |
| Length | 93 |
| Max. P | 0.910069 |

| Location | 13,702,861 – 13,702,954 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -14.80 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13702861 93 + 23771897 UACGCAGUUGUAACCCGGCCGCACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUACCCACAGC ...(((.(((......(((.............((((.(((((((.....))))))).))))...))).....)))..)))............. ( -14.89) >DroPse_CAF1 100103 88 + 1 AACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUC-AAGAUGCACACAUCCA---- ...(.(((((......(....)..........((((.(((((((.....))))))).)))).....))))))-..((((....))))..---- ( -14.00) >DroSec_CAF1 110077 93 + 1 UACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUACCCACAGA ...(((.(((......(((.............((((.(((((((.....))))))).))))...))).....)))..)))............. ( -14.89) >DroYak_CAF1 119785 93 + 1 UACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUCCAAGAUGCACAUACCCACAGC ...(((((((.....)))).............((((.(((((((.....))))))).))))................)))............. ( -13.20) >DroAna_CAF1 166334 92 + 1 UACGCAUUUGUAACCCGGCCGCACUUACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUAUACCCGG- ...((((((.......(((.............((((.(((((((.....))))))).))))...))).......))))))............- ( -17.83) >DroPer_CAF1 101545 88 + 1 AACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUC-AAGAUGCACACAUCCA---- ...(.(((((......(....)..........((((.(((((((.....))))))).)))).....))))))-..((((....))))..---- ( -14.00) >consensus UACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUCCAAGAUGCACAUACCCACAG_ ...(((((.((......)).))..........((((.(((((((.....))))))).))))................)))............. (-12.40 = -12.35 + -0.05)

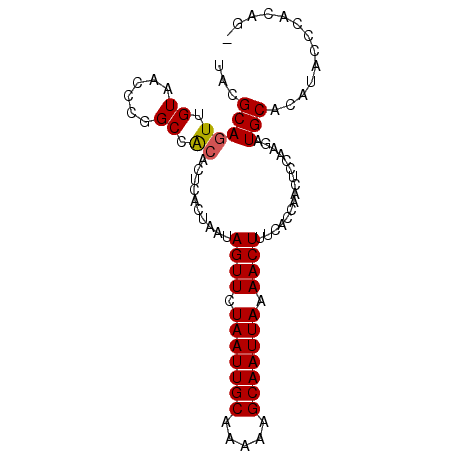

| Location | 13,702,861 – 13,702,954 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -22.00 |
| Energy contribution | -21.75 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13702861 93 - 23771897 GCUGUGGGUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGCGGCCGGGUUACAACUGCGUA ...(..(...((((...(((((.(((.((...((((((((((((.....))))))))))))....)).)..))..))))).)))).)..)... ( -25.80) >DroPse_CAF1 100103 88 - 1 ----UGGAUGUGUGCAUCUU-GAGUUGGUGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUU ----.(((((....))))).-.......(((.((((((((((((.....)))))))))))).)))(((..((((((...))))))..)))... ( -23.70) >DroSec_CAF1 110077 93 - 1 UCUGUGGGUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUA ...(..(...((((...(((((.(((.((...((((((((((((.....))))))))))))....)).)))....))))).)))).)..)... ( -25.00) >DroYak_CAF1 119785 93 - 1 GCUGUGGGUAUGUGCAUCUUGGAGUUGGUGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUA ...(..(...((((...((((((.(((.(((.((((((((((((.....)))))))))))).))).))).)....))))).)))).)..)... ( -25.20) >DroAna_CAF1 166334 92 - 1 -CCGGGUAUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUAAGUGCGGCCGGGUUACAAAUGCGUA -....((((.((((...(((((.((..((...((((((((((((.....))))))))))))....))....))..))))).)))).))))... ( -27.40) >DroPer_CAF1 101545 88 - 1 ----UGGAUGUGUGCAUCUU-GAGUUGGUGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUU ----.(((((....))))).-.......(((.((((((((((((.....)))))))))))).)))(((..((((((...))))))..)))... ( -23.70) >consensus _CUGUGGAUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUA .............(((.....((.(((((...((((((((((((.....))))))))))))(((....)))...))))).)).....)))... (-22.00 = -21.75 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:42 2006