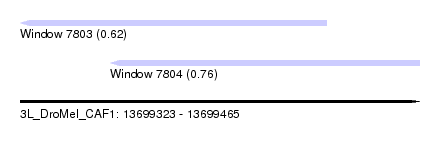
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,699,323 – 13,699,465 |
| Length | 142 |
| Max. P | 0.757949 |

| Location | 13,699,323 – 13,699,432 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.25 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.04 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
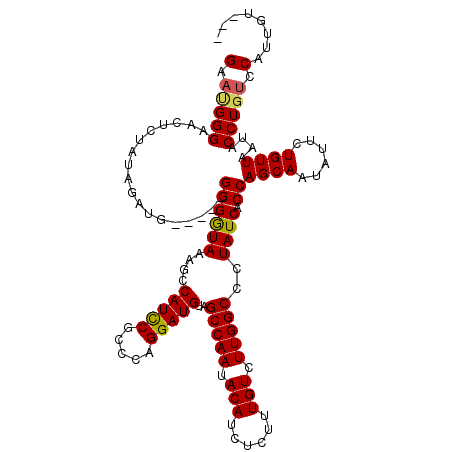
>3L_DroMel_CAF1 13699323 109 - 23771897 CCAACCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCUUAUCACCAGCAAUAUUCUGUUAAUACCUGUCCAUUGU---------UCGGUACACUCCUGCAGGGCACAAGUUAUUGG ((...((((((((.(((((.(((......))).)))))......(((((((((...(.((....))..)...)))))---------).)))))).)))))...))..(((....))). ( -25.00) >DroSec_CAF1 106685 109 - 1 CCGCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCUUAUCACCAGCAAUAUUCUGUUAAUACCUGUCCAUUGU---------UCGGUACACUCCUGCAGGGCACAAGUUAUUGA ..((((..((((..(((((.(((......))).)))))..))))(((((((((...(.((....))..)...)))))---------).)))...........)))).(((....))). ( -28.40) >DroEre_CAF1 112747 109 - 1 CCUCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCCUAUCACCAGCAAUAUUCUGUUAGUACCUGUCCAUUGU---------UCGGUAUAUUCCUGCAGGACACCAGUUAUUGA ........((((..(((((.(((......))).)))))..)))).....(((((..(((...((.(((((....(((---------.....))).....)))))))..))).))))). ( -24.00) >DroYak_CAF1 115892 118 - 1 UCGCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCCUAUCACCAGCAAUAUGCUGUUGGCACCUGCCCUUUGUACAGUUGUUUCGGUAUAUUCCUGCAGGACACAAGUUAUUGG ..(((...((((..(((((.(((......))).)))))..))))..((((.....))))..))).(((((....(((((.(......).))))).....)))))...(((....))). ( -33.70) >consensus CCGCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCCUAUCACCAGCAAUAUUCUGUUAAUACCUGUCCAUUGU_________UCGGUACACUCCUGCAGGACACAAGUUAUUGA ........((((..(((((.(((......))).)))))..)))).....(((((...(((.....(((((....(((..............))).....)))))..)))...))))). (-20.23 = -20.04 + -0.19)


| Location | 13,699,355 – 13,699,465 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.01 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -23.67 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13699355 110 - 23771897 GAAAGGGAACUCUAUAGAUG-----GGGAUAAAGCCAUCCAACCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCUUAUCACCAGCAAUAUUCUGUUAAUACCUGUCCAUUGU--- ((.(((.(((...(((..((-----(.(((((.((((..(((...(((((((.....)))))))..)))...)))).))))).)))....)))...)))....))).))......--- ( -29.20) >DroSec_CAF1 106717 110 - 1 GAACGGGAACUCUAUAGAUG-----GGGGUAAAGCCAUCCGCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCUUAUCACCAGCAAUAUUCUGUUAAUACCUGUCCAUUGU--- (.(((((......(((((.(-----((((.....)).)))((....((((..(((((.(((......))).)))))..))))....)).....))))).....))))).).....--- ( -29.70) >DroEre_CAF1 112779 115 - 1 GAAUGGGAACUCUAUAGCCGGGCGGGGGGUAAAUCCAUCCUCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCCUAUCACCAGCAAUAUUCUGUUAGUACCUGUCCAUUGU--- (.(((((.(((..((((....((((((((.........))))))..((((..(((((.(((......))).)))))..))))....))......)))).))).))))).).....--- ( -33.50) >DroYak_CAF1 115930 113 - 1 GAAUGGGCACUCCAUAGAUG-----GGGGUAAAUCCAUUCGCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCCUAUCACCAGCAAUAUGCUGUUGGCACCUGCCCUUUGUACA ....(((((..(((..((((-----((........(((((.....)))))..(((((.(((......))).)))))))))))..((((.....)))).)))....)))))........ ( -36.70) >consensus GAAUGGGAACUCUAUAGAUG_____GGGGUAAAGCCAUCCGCCCAGGAUGUAGCCAAUACAUCUCUUUGUCUUGGCCCUAUCACCAGCAAUAUUCUGUUAAUACCUGUCCAUUGU___ (.(((((..................((((((....(((((.....)))))..(((((.(((......))).)))))..)))).))((((......))))....))))).)........ (-23.67 = -23.68 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:35 2006