| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,391,507 – 1,391,613 |
| Length | 106 |
| Max. P | 0.989022 |

| Location | 1,391,507 – 1,391,613 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.03 |
| Mean single sequence MFE | -17.56 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1391507 106 + 23771897 C-------GGCCAAAACAUCUUUCAAUUAACGGGCCAUGACCGUGGCAAAGGCCAAAACCGAUACCACGGCCCAGUCCACCACUACCCAAACACAACCCAUACUUCAAAUCCC .-------((((....................)))).((.((((((....((......))....))))))..))....................................... ( -18.25) >DroPse_CAF1 72676 93 + 1 UGAGAGCCACACAAAACAUAUUUCAAUUAACAGGCCAUGACCGUGGCAAAGGCUAAGGCUAAGACCAACACCAA---CACCAGCACCGA-----------------GAGACCU ....((((...............(........)(((((....)))))...))))..((......))........---............-----------------....... ( -12.40) >DroSec_CAF1 46636 80 + 1 C-------GGCCAAAACAUCUUUCAAUUAACGGGCCAUGACCGUGGCAAAGGCCAAAACCGAUACCACGGCCCAUACUUC--------------------------AAGUCCC .-------((((....................))))(((.((((((....((......))....))))))..))).....--------------------------....... ( -19.25) >DroSim_CAF1 44087 80 + 1 C-------GGCCAAAACAUCUUUCAAUUAACGGGCCAUGACCGUGGCAAAGGCCAAAACCGAUACCACGGCCCAUACUUC--------------------------AAGUCCC .-------((((....................))))(((.((((((....((......))....))))))..))).....--------------------------....... ( -19.25) >DroEre_CAF1 48050 97 + 1 UGU-----GGCCCAAACAUAUUUCAAUUAACGGGCCAUGACCGUGGCAAAGGCCAAAACCGAUACCACAGCCCAGUCCACCAGCACC--AACACUUC---------GCACCCA .((-----(((((..................)))))))....((((....((......))....)))).((..(((...........--...)))..---------))..... ( -19.61) >DroYak_CAF1 49658 86 + 1 G-------GGCCAAAACAUAUUUCAAUUAACGGGCCAUGACCGUGGCAAAGGCCAAAACCGAUACCACAGUCCAGUCCACU---ACC--AACACU---------------CCA .-------.......................((((...(((.((((....((......))....)))).)))..))))...---...--......---------------... ( -16.60) >consensus C_______GGCCAAAACAUAUUUCAAUUAACGGGCCAUGACCGUGGCAAAGGCCAAAACCGAUACCACGGCCCAGUCCACC___ACC___________________AAGUCCC ........((((...........(........)(((((....)))))...))))........................................................... (-11.64 = -11.70 + 0.06)

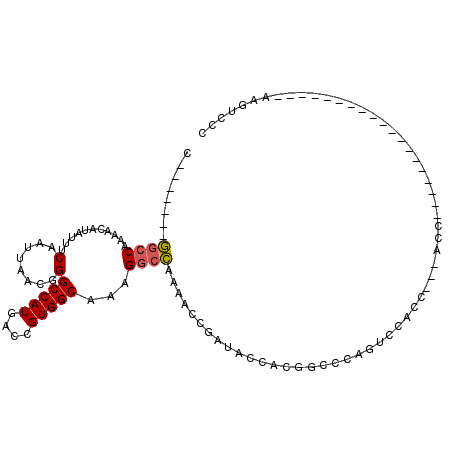
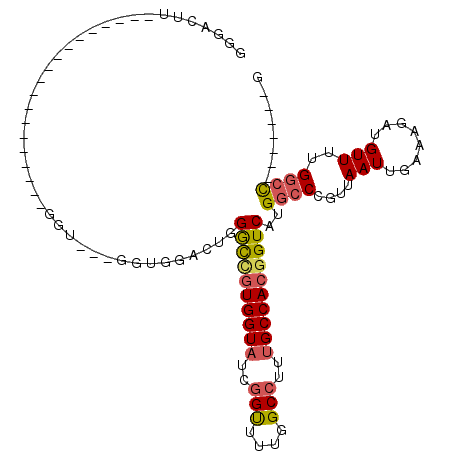
| Location | 1,391,507 – 1,391,613 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.03 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -21.55 |
| Energy contribution | -21.83 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1391507 106 - 23771897 GGGAUUUGAAGUAUGGGUUGUGUUUGGGUAGUGGUGGACUGGGCCGUGGUAUCGGUUUUGGCCUUUGCCACGGUCAUGGCCCGUUAAUUGAAAGAUGUUUUGGCC-------G .((.(((.((..((((((..(.......((((.....))))((((((((((..(((....)))..)))))))))))..))))))...)).)))..........))-------. ( -35.60) >DroPse_CAF1 72676 93 - 1 AGGUCUC-----------------UCGGUGCUGGUG---UUGGUGUUGGUCUUAGCCUUAGCCUUUGCCACGGUCAUGGCCUGUUAAUUGAAAUAUGUUUUGUGUGGCUCUCA .((((.(-----------------.(((.(((((((---..(((...(((....)))...)))..))))).(((....)))...............)).))).).)))).... ( -22.20) >DroSec_CAF1 46636 80 - 1 GGGACUU--------------------------GAAGUAUGGGCCGUGGUAUCGGUUUUGGCCUUUGCCACGGUCAUGGCCCGUUAAUUGAAAGAUGUUUUGGCC-------G .......--------------------------........((((((((((..(((....)))..))))))))))..((((.....(((....))).....))))-------. ( -31.50) >DroSim_CAF1 44087 80 - 1 GGGACUU--------------------------GAAGUAUGGGCCGUGGUAUCGGUUUUGGCCUUUGCCACGGUCAUGGCCCGUUAAUUGAAAGAUGUUUUGGCC-------G .......--------------------------........((((((((((..(((....)))..))))))))))..((((.....(((....))).....))))-------. ( -31.50) >DroEre_CAF1 48050 97 - 1 UGGGUGC---------GAAGUGUU--GGUGCUGGUGGACUGGGCUGUGGUAUCGGUUUUGGCCUUUGCCACGGUCAUGGCCCGUUAAUUGAAAUAUGUUUGGGCC-----ACA ..(((.(---------(.(((...--...)))..)).))).((((((((((..(((....)))..)))))))))).(((((((...((......))...))))))-----).. ( -37.40) >DroYak_CAF1 49658 86 - 1 UGG---------------AGUGUU--GGU---AGUGGACUGGACUGUGGUAUCGGUUUUGGCCUUUGCCACGGUCAUGGCCCGUUAAUUGAAAUAUGUUUUGGCC-------C ...---------------.(((((--..(---(((((.(((((((((((((..(((....)))..)))))))))).))).)))))).....))))).........-------. ( -26.30) >consensus GGGACUU___________________GGU___GGUGGACUGGGCCGUGGUAUCGGUUUUGGCCUUUGCCACGGUCAUGGCCCGUUAAUUGAAAGAUGUUUUGGCC_______G .........................................((((((((((..(((....)))..))))))))))..((((....(((........)))..))))........ (-21.55 = -21.83 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:28 2006