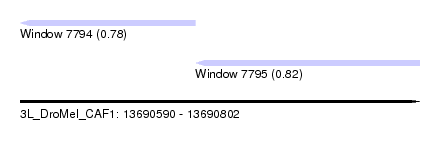
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,690,590 – 13,690,802 |
| Length | 212 |
| Max. P | 0.820327 |

| Location | 13,690,590 – 13,690,683 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -25.18 |
| Energy contribution | -24.56 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13690590 93 - 23771897 CUUCACCAUCAACAUCCUUCUUCAGCCUUUCGCUCGCAUUGACACGCAUGUUGCAGCCGGCCUGCAACACAGUGUGCGAAGGUCCUGGCACGA .....................((((.(((((((..((((((.......((((((((.....)))))))))))))))))))))..))))..... ( -26.11) >DroSec_CAF1 97940 87 - 1 CUUCGC-----ACA-CCAUCUUCAGCCUUUCGCUCGCAUUGACACGCAUGUUGCAGCCGGCCUGCAACACAGUGUGCGAAGGUCCUGGCACGA ((((((-----(((-(.......(((.....))).((........)).((((((((.....))))))))..))))))))))((....)).... ( -29.70) >DroEre_CAF1 103604 87 - 1 CUUCAC-----GCU-GCCUCCUCGGCCUUUCGCUCGCAUUGACACGCAUGUUGCAGCCGGCCUGCAAUACAGUGUGCGAAGGUCCUGGCACGA .....(-----(.(-(((.....((((((.(((..((((((.......((((((((.....)))))))))))))))))))))))..)))))). ( -31.31) >DroYak_CAF1 107030 77 - 1 ----------------CUUCCUUGGCCUUUCGCUCGCAUUGACACGCAUGUUGCAGCCGGCCUGCAACACAGUGUGCGAAGGUCCUGGCACGA ----------------...((..((((((.(((..((((((.......((((((((.....)))))))))))))))))))))))..))..... ( -28.31) >consensus CUUCAC_____ACA_CCUUCCUCAGCCUUUCGCUCGCAUUGACACGCAUGUUGCAGCCGGCCUGCAACACAGUGUGCGAAGGUCCUGGCACGA .....................((((.(((((((..((((((.......((((((((.....)))))))))))))))))))))..))))..... (-25.18 = -24.56 + -0.62)

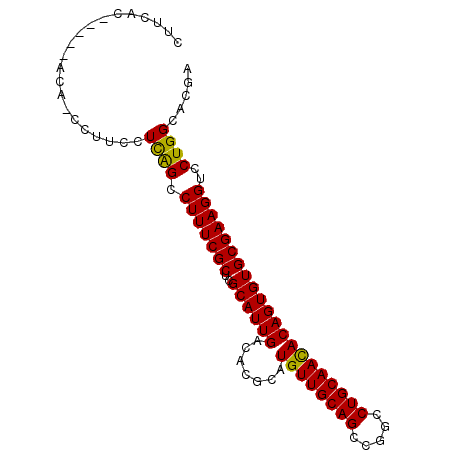

| Location | 13,690,683 – 13,690,802 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13690683 119 - 23771897 AAUGUGUCGGUGAUUUACAACAAUGUCGCCUACACAAAUUUUCCAAAUGUGUUUUUGCCACGAAAGGAAACCUAGGCC-AGUCGUCGCACACACAGGACCUACCUUCUACCCGCUGCCCC ..(((((.((((((..........)))))).))))).....(((...(((((...(((.((((..((.........))-..)))).))).))))))))...................... ( -28.90) >DroPse_CAF1 86938 98 - 1 AAUGUGUCGGUGAUUUACAACAAUGUCGCCUACACAAAUUUUCCAAAUGUGUUUUUGCCACGAAAGCAAACGCAGGCC-AGGC--CGCA---A------CACCCACUUCA---------- ..(((((.((((((..........)))))).)))))............((((((((((.......))))).((.(((.-..))--))))---)------)))........---------- ( -24.50) >DroEre_CAF1 103691 119 - 1 AAUGUGUCGGUGAUUUACAACAAUGUCGCCUACACAAAUUUUCCAAAUGUGUUUUUGCCACGAAAGCAAACCUAGGCC-AGUCGUCGCACACACAGGACCUACCUCCCUCCCGCUGACCC ..(((((.((((((..........)))))).))))).....(((...(((((...(((.((((..((........)).-..)))).))).))))))))...................... ( -28.80) >DroYak_CAF1 107107 109 - 1 AAUGUGUCGGUGAUUUACAACAAUGUCGCCUACACAAAUUUUCCAAAUGUGUUUUUGCCACGAAAGCAAACCUAGGCC-AGUCGUCGCACACACAGGACCAACCACCUCC---------- ..(((((.((((((..........)))))).))))).....(((...(((((...(((.((((..((........)).-..)))).))).))))))))............---------- ( -28.80) >DroAna_CAF1 155907 112 - 1 AAUGUGUCGGUGAUUAACAACAAUGUCGCCUACACAAAUUUUCCAAAUGCGUUUUUGCCACGAAAGCAAGCGCAGGCCUAGGCCUGGUCCCAACAAAACGCACCCACU-------GGCC- ..(((((.((((((..........)))))).)))))......(((..((((((((.((..(....)...)).(((((....)))))........)))))))).....)-------))..- ( -34.40) >DroPer_CAF1 88176 98 - 1 AAUGUGUCGGUGAUUUACAACAAUGUCGCCUACACAAAUUUUCCAAAUGUGUUUUUGCCACGAAAGCAAACGCAGGCC-AGGC--CGCA---A------CACCCACUUCA---------- ..(((((.((((((..........)))))).)))))............((((((((((.......))))).((.(((.-..))--))))---)------)))........---------- ( -24.50) >consensus AAUGUGUCGGUGAUUUACAACAAUGUCGCCUACACAAAUUUUCCAAAUGUGUUUUUGCCACGAAAGCAAACCCAGGCC_AGGCGUCGCACACACAGGACCAACCACCUCC__________ ..(((((.((((((..........)))))).)))))......((.....((((((((...))))))))......))............................................ (-16.83 = -17.17 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:27 2006