
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,686,728 – 13,686,827 |
| Length | 99 |
| Max. P | 0.571388 |

| Location | 13,686,728 – 13,686,827 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -16.65 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13686728 99 + 23771897 ------------ACUCUCUCCCGCUGUCCGAGCUGGAAAACUUUUCUUUUAGCCGGGUUUUUCCUUCUUUGC--UGCGGUUGACAGCC-------AACAGCGACAACAUUUUAAAGUCGC ------------.((.((..((((.(((((.(((((((........))))))))))..............))--.))))..)).))..-------....(((((...........))))) ( -23.83) >DroPse_CAF1 82400 112 + 1 CUACUUCUCUU-UCUCUUUUCUGUUGUCCGAGCAUACAAAAUUUUCUUUUAGCCGAGUUUUUCCCACUUGGCUCUGCGGUUGACAGCC-------AACAGCGACAACAUUUUAAGGCCGC ...........-..((((...(((((((......................((((((((.......))))))))..((.((((.....)-------))).)))))))))....)))).... ( -25.20) >DroSec_CAF1 94036 99 + 1 ------------ACUCUCUCCCGCUGUCCGAGCUGGAACACUUUUCUUUUAGCCGGGUUCUUCCUUCUUUGC--UGCGGUUGACAGCC-------AACAGCGACAACAUUUUAAAGUCGC ------------.((.((..((((.(((((.(((((((........))))))))))..............))--.))))..)).))..-------....(((((...........))))) ( -23.53) >DroEre_CAF1 99757 99 + 1 ------------ACUCUCGCCCGCUGUCCGAGCUGAAAAACUUUUCUUUUAGCCGGGUUUUUCCUUCGUUGC--UGCGGUUGACAGCC-------AACAGCGACAACAUUUUAAAGUCGU ------------.........((((.((((.(((((((........)))))))))))..........(((((--((.(((.....)))-------..)))))))..........)).)). ( -25.40) >DroYak_CAF1 103164 104 + 1 --------------UCUCUGCCGCUGUCCGAGCUGAAAAACUUUUCUUUUAGCCGGGUUUUUCCUUCUUGGC--UGCGCUUGACAGCCAACAGCCAACAGCGACAACAUUUUAAAGUCGC --------------....((.(((((((((.(((((((........))))))))))............((((--((.(((....)))...)))))))))))).))............... ( -31.50) >DroPer_CAF1 83705 113 + 1 GUACUUCUCUUUUCUCUUUUCUGUUGUCCGAGCAUACAAAAUUUUCUUUUAGCCGAGUUUUUCCCACUUGGCUCUGCGGUUGACAGCC-------AACAGCGACAACAUUUUAAGGCCGC ..............((((...(((((((......................((((((((.......))))))))..((.((((.....)-------))).)))))))))....)))).... ( -25.20) >consensus ____________ACUCUCUCCCGCUGUCCGAGCUGAAAAACUUUUCUUUUAGCCGGGUUUUUCCUUCUUGGC__UGCGGUUGACAGCC_______AACAGCGACAACAUUUUAAAGUCGC ......................((((((((.(((((((........)))))))))...............((...))....))))))............(((((...........))))) (-16.65 = -17.40 + 0.75)
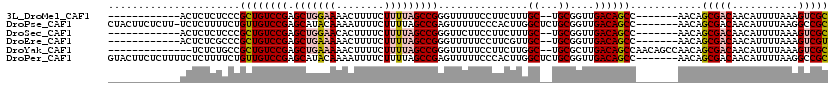

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:23 2006