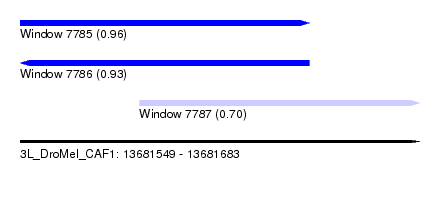
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,681,549 – 13,681,683 |
| Length | 134 |
| Max. P | 0.964153 |

| Location | 13,681,549 – 13,681,646 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -24.75 |
| Energy contribution | -27.78 |
| Covariance contribution | 3.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
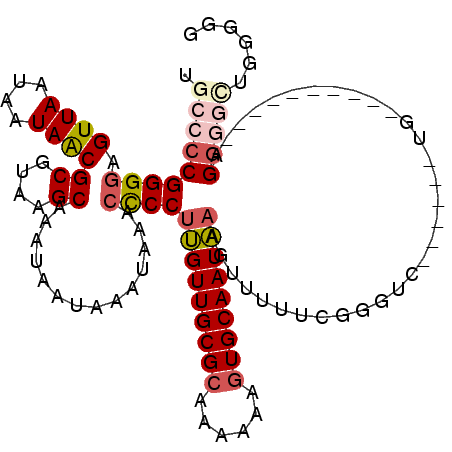
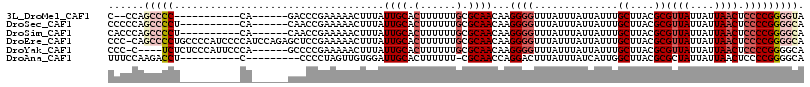
>3L_DroMel_CAF1 13681549 97 + 23771897 UACCCCGGGGAGUUAAUAAUAACGCGUAAGCAAAUAAUAAAUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGGUC------UG-----------GGGGCUGG--G ..((((((((.((((....))))((....))..............))))(((((((((......))))))))).............------.)-----------))).....--. ( -32.30) >DroSec_CAF1 88798 100 + 1 UGCCCCGGGGAGUUAAUAAUAACGCGUAAGCAAAUAAUAAAUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGUUG------UG----------AGGGGCUGGGGG ..((((((((.((((....))))((....))..............))))(((((((((......)))))))))(((((((((....------))----------))))))))))). ( -37.30) >DroSim_CAF1 99410 100 + 1 UGCCCCGGGGAGUUAAUAAUAACGCGUAAGCAAAUAAUAAAUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGUUG------UG----------AGGGGCUGGGUG .(((((((((.((((....))))((....))..............))))(((((((((......))))))))).............------..----------.)))))...... ( -35.30) >DroEre_CAF1 94205 115 + 1 UGCCCCGGGGAGUUAAUAAUAACGCGUAAGCAAAUAAUAAAUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGAGCUCUGGAUGGGGAUGGGGCAGGGGCUG-GGG .(((((((((.((((....))))((....))..............))))(((((((((......)))))))))...........((((((........)))))).)))))..-... ( -40.90) >DroYak_CAF1 97852 105 + 1 UGCCCCGGGGAGUUAAUAAUAACGCGUAAGCAAAUAAUAAAUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGGGC------UGGGAAUGGGAGAGA----G-GGG .(((((((((.((((....))))((....))..............))))(((((((((......)))))))))........)))))------...............----.-... ( -35.40) >DroAna_CAF1 134205 96 + 1 UGCCCCGGGGAGUUAAUAAUAGCGCGUAAGCCAAUGAUAAAUAAAGUCCUGGUUGCG-AAAAAAGUGCAAUCCACAACUAGGGG---------G----------AGGUCUUGGAAA ..((((.((((((((....))))((....))...............))))(((((((-.......)))))))........))))---------.----------............ ( -23.00) >consensus UGCCCCGGGGAGUUAAUAAUAACGCGUAAGCAAAUAAUAAAUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGGUC______UG__________AGGGGCUGGGGG .(((((((((.((((....))))((....))..............))))(((((((((......)))))))))................................)))))...... (-24.75 = -27.78 + 3.03)

| Location | 13,681,549 – 13,681,646 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -17.14 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13681549 97 - 23771897 C--CCAGCCCC-----------CA------GACCCGAAAAACUUUAUUGCACUUUUUUGCGCAACAAGGGGUUUAUUUAUUAUUUGCUUACGCGUUAUUAUUAACUCCCCGGGGUA .--...(((((-----------..------................((((.(......).))))...((((..............((....))((((....)))).))))))))). ( -23.30) >DroSec_CAF1 88798 100 - 1 CCCCCAGCCCCU----------CA------CAACCGAAAAACUUUAUUGCACUUUUUUGCGCAACAAGGGGUUUAUUUAUUAUUUGCUUACGCGUUAUUAUUAACUCCCCGGGGCA ......(((((.----------..------................((((.(......).))))...((((..............((....))((((....)))).))))))))). ( -26.00) >DroSim_CAF1 99410 100 - 1 CACCCAGCCCCU----------CA------CAACCGAAAAACUUUAUUGCACUUUUUUGCGCAACAAGGGGUUUAUUUAUUAUUUGCUUACGCGUUAUUAUUAACUCCCCGGGGCA ......(((((.----------..------................((((.(......).))))...((((..............((....))((((....)))).))))))))). ( -26.00) >DroEre_CAF1 94205 115 - 1 CCC-CAGCCCCUGCCCCAUCCCCAUCCAGAGCUCCGAAAAACUUUAUUGCACUUUUUUGCGCAACAAGGGGUUUAUUUAUUAUUUGCUUACGCGUUAUUAUUAACUCCCCGGGGCA ...-........(((((.............................((((.(......).))))...((((..............((....))((((....)))).))))))))). ( -26.30) >DroYak_CAF1 97852 105 - 1 CCC-C----UCUCUCCCAUUCCCA------GCCCCGAAAAACUUUAUUGCACUUUUUUGCGCAACAAGGGGUUUAUUUAUUAUUUGCUUACGCGUUAUUAUUAACUCCCCGGGGCA ...-.----...............------(((((...........((((.(......).))))...((((..............((....))((((....)))).))))))))). ( -26.50) >DroAna_CAF1 134205 96 - 1 UUUCCAAGACCU----------C---------CCCCUAGUUGUGGAUUGCACUUUUUU-CGCAACCAGGACUUUAUUUAUCAUUGGCUUACGCGCUAUUAUUAACUCCCCGGGGCA ............----------.---------((((.((((.(((.((((........-.))))))).))))...........((((......)))).............)))).. ( -21.40) >consensus CCCCCAGCCCCU__________CA______GACCCGAAAAACUUUAUUGCACUUUUUUGCGCAACAAGGGGUUUAUUUAUUAUUUGCUUACGCGUUAUUAUUAACUCCCCGGGGCA ......(((((...................................((((.(......).))))...((((..............((....))((((....)))).))))))))). (-17.14 = -18.58 + 1.44)
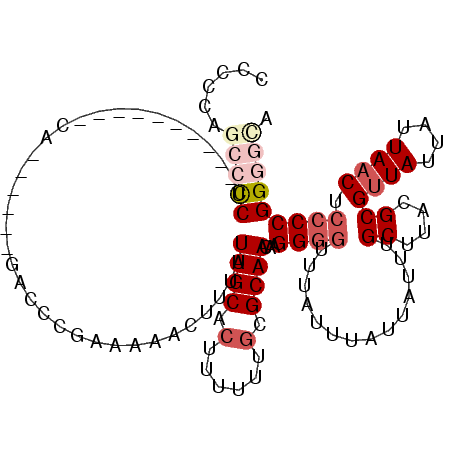
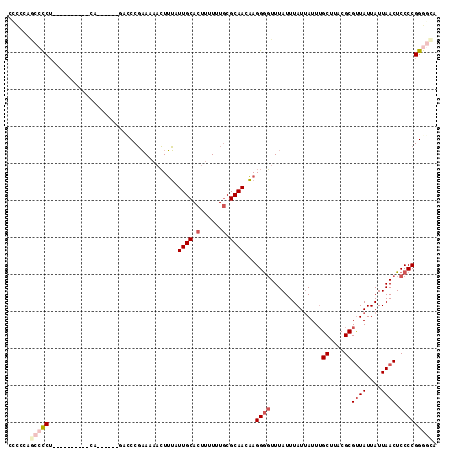
| Location | 13,681,589 – 13,681,683 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.37 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
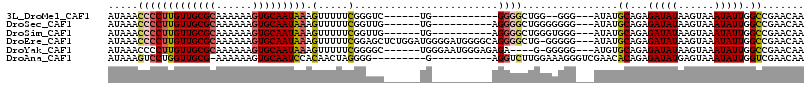
>3L_DroMel_CAF1 13681589 94 + 23771897 AUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGGUC------UG-----------GGGGCUGG--GGG---AUAUGCAGAGAUAUAAGUAAAUAUUGGCCGAACAA (((..(((((((((((((......))))))))).........((((------..-----------..))))))--)).---.)))((...(((((......))))).))....... ( -24.90) >DroSec_CAF1 88838 97 + 1 AUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGUUG------UG----------AGGGGCUGGGGGGG---AUAUGCAGAGAUAUAAGUAAAUAUUGGCCGAACAA (((..(((((((((((((......)))))))).(((((((((....------))----------))))))).))))).---.)))((...(((((......))))).))....... ( -27.80) >DroSim_CAF1 99450 97 + 1 AUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGUUG------UG----------AGGGGCUGGGUGGG---AUAUGCAGAGAUAUAAGUAAAUAUUGGCCGAACAA ....((((.(((((((((......)))))))))(((((((((....------))----------)))))))))))...---....((...(((((......))))).))....... ( -24.20) >DroEre_CAF1 94245 112 + 1 AUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGAGCUCUGGAUGGGGAUGGGGCAGGGGCUG-GGGGG---AUAUGCAGAGAUAUAAGUAAAUAUUGGCCGAACAA (((..(((((((((((((......)))))))))(((((((....((((((........))))))))))))).-)))).---.)))((...(((((......))))).))....... ( -29.20) >DroYak_CAF1 97892 102 + 1 AUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGGGC------UGGGAAUGGGAGAGA----G-GGGGG---AUGUGCAGAGAUAUAAGUAAAUAUUGGCCGAACAA .....(((((((((((((......)))))))....(((..((....------......))..)))))----)-)))(.---.((.((...(((((......))))).))))..).. ( -25.50) >DroAna_CAF1 134245 96 + 1 AUAAAGUCCUGGUUGCG-AAAAAAGUGCAAUCCACAACUAGGGG---------G----------AGGUCUUGGAAAGGGUCGAACACAGAGAUAUGAGUAAAUAUUGGUCGAACAA ......(((((((((.(-..............).))))))))).---------.----------.(..(((....)))..)....((...(((((......))))).))....... ( -18.44) >consensus AUAAACCCCUUGUUGCGCAAAAAAGUGCAAUAAAGUUUUUCGGGUC______UG__________AGGGGCUGGGGGGG___AUAUGCAGAGAUAUAAGUAAAUAUUGGCCGAACAA .....(((((((((((((......))))))))).(.....)........................))))................((...(((((......))))).))....... (-14.42 = -14.37 + -0.05)
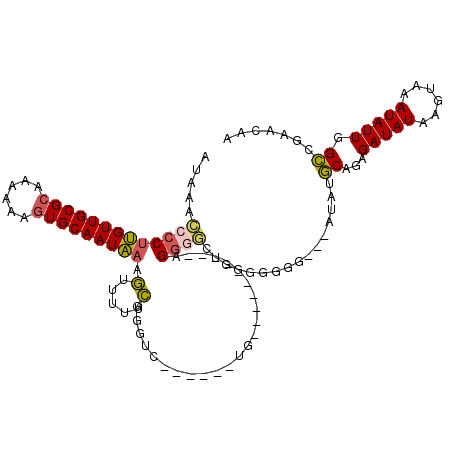
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:19 2006