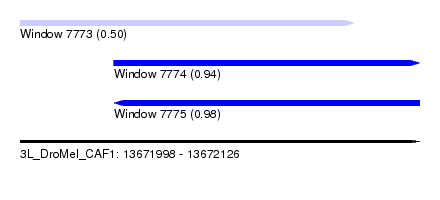
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,671,998 – 13,672,126 |
| Length | 128 |
| Max. P | 0.983908 |

| Location | 13,671,998 – 13,672,105 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.94 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -27.56 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13671998 107 + 23771897 UGAAAUGAAUGUGAUUGCCAUGCCU----------CCAUCCAUACGGCCAACACAGCUGCUGUUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCC-GCUU ......(((((((.((((((((((.----------..........)))......(((((((......(((......))).......)))))))...))))))))))))))...-.... ( -32.02) >DroPse_CAF1 66026 107 + 1 UGAAAUGAAUGUGAUUGCCA-GCGUGGCGACGCAUGCCACCGCCUGGCCCCC----------CCCCUUCCACUCAUAGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUCCCAGCUC ......(((((((...((((-((((((((.....))))).)).)))))....----------...((.(((...(((((((.(....))))))))..))).)))))))))........ ( -33.10) >DroSec_CAF1 79344 107 + 1 UGAAAUGAAUGUGAUUGCCAUGCCU----------CCAUCCACACGGCCAACACAGCUGCUGUUUCUGCCUCUAGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCC-GCUU ......(((((((.((((((((((.----------..........)))......(((((((......(((......))).......)))))))...))))))))))))))...-.... ( -32.02) >DroSim_CAF1 89522 107 + 1 UGAAAUGAAUGUGAUUGCCGUGCCU----------CCAUCCACACGGCCAACACAGCUGCUGUUUCUGCCUCUAGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCC-GCUU .........((((...((((((...----------.......))))))...))))((.((.((..(((((..(((((((((.(....)))))))))).)))))...))..)).-)).. ( -37.00) >DroEre_CAF1 84581 107 + 1 UGAAAUGAAUGUGAUUGCCAUGCCU----------CCGUCCACACGGCCAACACAGCUGCUGCUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCC-GCUU ......(((((((.((((((((...----------((((....))))...........((((((((((((......)))....)))))))))...)))))))))))))))...-.... ( -35.60) >DroYak_CAF1 87934 110 + 1 UGAAAUGAAUGUGAUUGCCAUGCCU-------CCUCCAUCCACACGGCCAACACAGCUGCUGUUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCC-GCUU ......(((((((.(((((((((.(-------((..((.((((..(((....((((...))))....)))....)))).))..))).))).......)))))))))))))...-.... ( -32.71) >consensus UGAAAUGAAUGUGAUUGCCAUGCCU__________CCAUCCACACGGCCAACACAGCUGCUGUUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCC_GCUU ......(((((((.((((((((.......................(((....((((...))))....)))....(((((((.(....)))))))))))))))))))))))........ (-27.56 = -27.93 + 0.38)



| Location | 13,672,028 – 13,672,126 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -25.88 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13672028 98 + 23771897 CAUACGGCCAACACAGCUGCUGUUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCCGCUUUUGGGGGCAUUCGAAAUUCCC ......(((..((.(((.((.((..(((((....(((((((.(....))))))))...)))))...))..)).)))..))..)))............. ( -31.00) >DroSec_CAF1 79374 98 + 1 CACACGGCCAACACAGCUGCUGUUUCUGCCUCUAGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCCGCUUUUUGGGGCAUUCAAAAUUCCC .(((((((.......)))).)))..(((((..(((((((((.(....)))))))))).)))))......((((.(.....).))))............ ( -31.40) >DroSim_CAF1 89552 98 + 1 CACACGGCCAACACAGCUGCUGUUUCUGCCUCUAGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCCGCUUUUUGGGGCAUUCAAAAUUCCC .(((((((.......)))).)))..(((((..(((((((((.(....)))))))))).)))))......((((.(.....).))))............ ( -31.40) >DroEre_CAF1 84611 98 + 1 CACACGGCCAACACAGCUGCUGCUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCCGCUUUUUGGCGCUUUCGAAAUUCCC ......(((((...(((.((((((((((((......)))....)))))))))......(((((.....))))))))..)))))............... ( -32.40) >DroYak_CAF1 87967 98 + 1 CACACGGCCAACACAGCUGCUGUUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCCGCUUUUUGGCACUCUCGUAAUUCCC ......(((((...(((.((.((..(((((....(((((((.(....))))))))...)))))...))..)).)))..)))))............... ( -30.30) >consensus CACACGGCCAACACAGCUGCUGUUUCUGCCUCUCGUGGCUGUCGGAAGCAGUUAUUAUGGCAGCAUAUUUGCCGCUUUUUGGGGCAUUCGAAAUUCCC .......((((...(((.((((((((((((......)))....)))))))))......(((((.....))))))))..))))................ (-25.88 = -25.92 + 0.04)

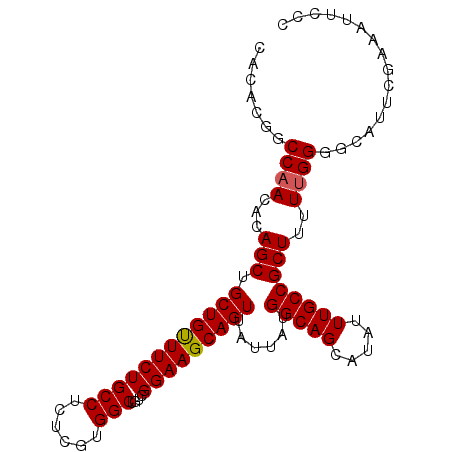
| Location | 13,672,028 – 13,672,126 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.54 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13672028 98 - 23771897 GGGAAUUUCGAAUGCCCCCAAAAGCGGCAAAUAUGCUGCCAUAAUAACUGCUUCCGACAGCCACGAGAGGCAGAAACAGCAGCUGUGUUGGCCGUAUG (((.............)))....(((((.((((((((((........(((((((((.......)).))))))).....))))).))))).)))))... ( -32.84) >DroSec_CAF1 79374 98 - 1 GGGAAUUUUGAAUGCCCCAAAAAGCGGCAAAUAUGCUGCCAUAAUAACUGCUUCCGACAGCCACUAGAGGCAGAAACAGCAGCUGUGUUGGCCGUGUG (((............))).....(((((.((((((((((........(((((((............))))))).....))))).))))).)))))... ( -30.12) >DroSim_CAF1 89552 98 - 1 GGGAAUUUUGAAUGCCCCAAAAAGCGGCAAAUAUGCUGCCAUAAUAACUGCUUCCGACAGCCACUAGAGGCAGAAACAGCAGCUGUGUUGGCCGUGUG (((............))).....(((((.((((((((((........(((((((............))))))).....))))).))))).)))))... ( -30.12) >DroEre_CAF1 84611 98 - 1 GGGAAUUUCGAAAGCGCCAAAAAGCGGCAAAUAUGCUGCCAUAAUAACUGCUUCCGACAGCCACGAGAGGCAGAAGCAGCAGCUGUGUUGGCCGUGUG .((.....(....)..)).....(((((.((((((((((........(((((((((.......)).))))))).....))))).))))).)))))... ( -31.82) >DroYak_CAF1 87967 98 - 1 GGGAAUUACGAGAGUGCCAAAAAGCGGCAAAUAUGCUGCCAUAAUAACUGCUUCCGACAGCCACGAGAGGCAGAAACAGCAGCUGUGUUGGCCGUGUG .((...(((....))))).....(((((.((((((((((........(((((((((.......)).))))))).....))))).))))).)))))... ( -32.92) >consensus GGGAAUUUCGAAUGCCCCAAAAAGCGGCAAAUAUGCUGCCAUAAUAACUGCUUCCGACAGCCACGAGAGGCAGAAACAGCAGCUGUGUUGGCCGUGUG (((............))).....(((((.((((((((((........(((((((((.......)).))))))).....))))).))))).)))))... (-27.74 = -28.54 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:07 2006