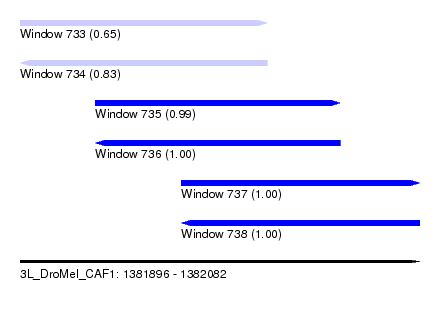
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,381,896 – 1,382,082 |
| Length | 186 |
| Max. P | 0.999941 |

| Location | 1,381,896 – 1,382,011 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381896 115 + 23771897 AUCAAUUAUCGUUAACCAACACUGA--AUGCAAGAAUGUCAUAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAU .........................--(((((....)).))).(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))... ( -27.70) >DroSim_CAF1 34652 114 + 1 AUU-AUAAUCGUUAACCAACACUAA--AUCAGAAAAUGCCAAAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAU ...-.....................--................(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))... ( -27.60) >DroEre_CAF1 38423 114 + 1 AUC-CUAAUCGUUAGUCAACACUGA--AUCCAGGGAUGUCAACUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAU (((-(((((((((((.(((...((.--...))((((.(....)))))...))).)))))))))))))).......(((((.((....)).(((((.......)))))..)))))... ( -38.20) >DroAna_CAF1 38648 116 + 1 AUC-UUAGCCGUUUACCAACACUAACUAUCGGGGGACGUUAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAC ...-...((((..((((..(((((((......(((((.....))))).....)).)))))....)).))..))))((..((((((.....))))))..))(((((.....))))).. ( -42.20) >consensus AUC_AUAAUCGUUAACCAACACUAA__AUCCAAGAAUGUCAAAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAU ...........................................(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))... (-27.52 = -27.53 + 0.00)

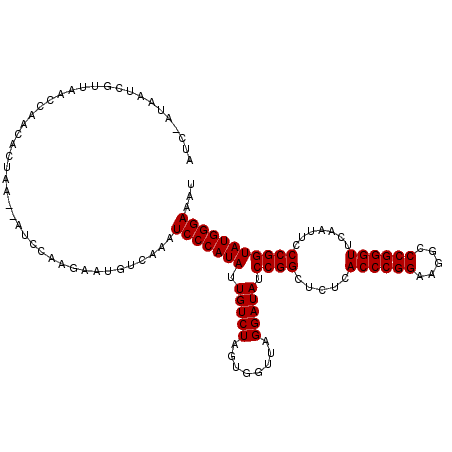
| Location | 1,381,896 – 1,382,011 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -36.11 |
| Consensus MFE | -30.19 |
| Energy contribution | -29.62 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381896 115 - 23771897 AUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUAUGACAUUCUUGCAU--UCAGUGUUGGUUAACGAUAAUUGAU ...(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))).(((.((....)))))--(((((((((.....)))).))))). ( -33.30) >DroSim_CAF1 34652 114 - 1 AUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUUUGGCAUUUUCUGAU--UUAGUGUUGGUUAACGAUUAU-AAU ...(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))).(..(((((.......--..)))))..)...........-... ( -32.40) >DroEre_CAF1 38423 114 - 1 AUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGUUGACAUCCCUGGAU--UCAGUGUUGACUAACGAUUAG-GAU ..........((((...((..((((((.....))))))..)).))))..(((((((.(..(((.((((((..(((((.(......).)))--)).)))))).)))..).))))-))) ( -35.60) >DroAna_CAF1 38648 116 - 1 GUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUAACGUCCCCCGAUAGUUAGUGUUGGUAAACGGCUAA-GAU ..(((((.....)))))((..((((((.....))))))..))((((..((.((....((((((.......(((((.....))))).......))))))..))))..))))...-... ( -43.14) >consensus AUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUUUGACAUCCCCGGAU__UCAGUGUUGGUUAACGAUUAU_GAU ...(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))).......................((((.....))))....... (-30.19 = -29.62 + -0.56)

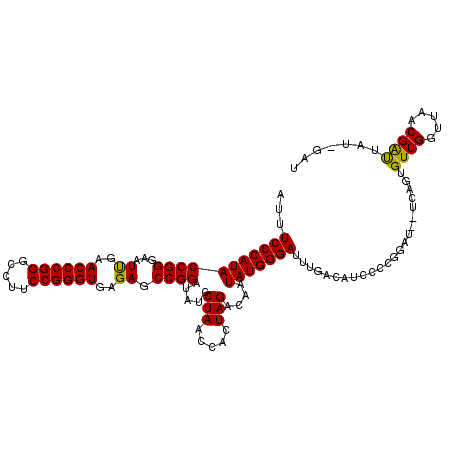
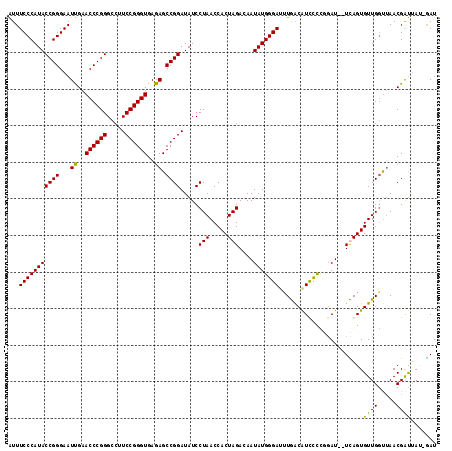
| Location | 1,381,931 – 1,382,045 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381931 114 + 23771897 GUCAUAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUGUGGUUGAAUUGUUUUUUCUUUUUCGAA---AUUAUU .((((((((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))..)))))(((((..(......)...))))).---...... ( -30.50) >DroEre_CAF1 38457 94 + 1 GUCAACUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUGUGGGUGCAUUCCU----------------------- .....(.((((((((..(((((((...((....)).....))))((....)).(((((.......)))))..)))..)))))))).).......----------------------- ( -29.00) >DroAna_CAF1 38684 100 + 1 GUUAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAC----------UCUUUUU------UCAAAAAAAUAAU- ......(((((((.(((((........))))).((((.((..((((((.....))))))..))...)))))))))))...----------.......------.............- ( -28.80) >consensus GUCAAAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUGUGG_UG_AUUCUUUUU______UC_AA___AU_AU_ ......(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))........................................ (-27.50 = -27.50 + 0.00)

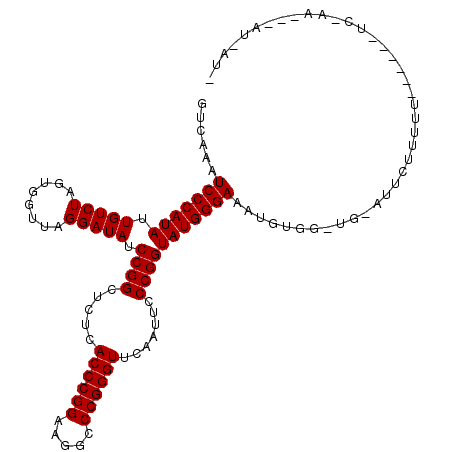
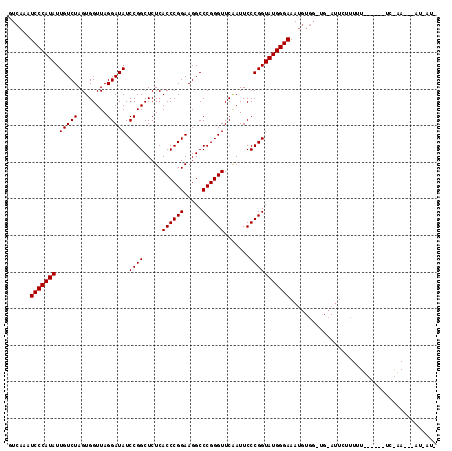
| Location | 1,381,931 – 1,382,045 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.70 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381931 114 - 23771897 AAUAAU---UUCGAAAAAGAAAAAACAAUUCAACCACAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUAUGAC .....(---(((......))))..............(((.(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))).))).. ( -32.80) >DroEre_CAF1 38457 94 - 1 -----------------------AGGAAUGCACCCACAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGUUGAC -----------------------.((.......)).((.((((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))).)).. ( -31.30) >DroAna_CAF1 38684 100 - 1 -AUUAUUUUUUUGA------AAAAAGA----------GUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUAAC -.....((((((..------..)))))----------)..(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))...... ( -31.80) >consensus _AU_AU___UU_GA______AAAAAGAAU_CA_CCACAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGAC ....................................((..(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))..)).. (-29.11 = -29.00 + -0.11)

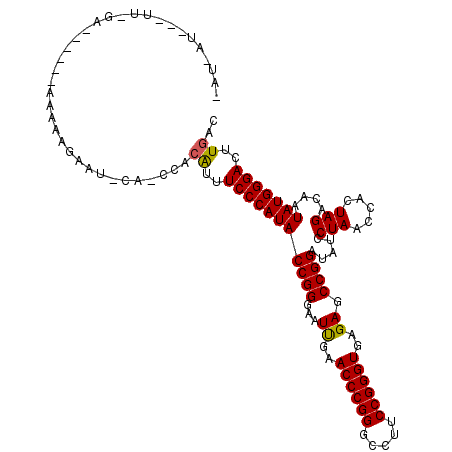
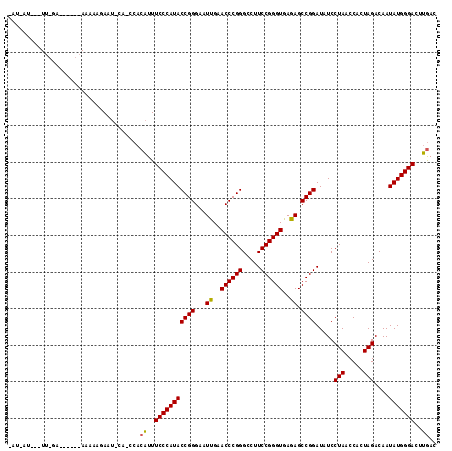
| Location | 1,381,971 – 1,382,082 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.94 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381971 111 + 23771897 UCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUGUGGUUGAAUUGUUUUUUCUUUUUCGAA---AUUAUUUUAAAAAUUCAAUGUUGUACAAAAUAUAAUCUCUAUG ..((((((.....)))))).((.(((((.....))))).))..((((((((((..((((......)))---)..))......))))))))((((((.....))))))....... ( -22.80) >DroEre_CAF1 38497 89 + 1 UCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUGUGGGUGCAUUCCU-------------------------UGUAAUACAAUGUAAUACAAAAUAUAAUUUCUCUG ....((....)).(((((.......)))))...(((((((..(((.....)))(-------------------------((((.(((...))).)))))......))))))).. ( -22.90) >DroAna_CAF1 38724 89 + 1 UCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAAC----------UCUUUUU------UCAAAAAAAUAAU-UUGCAAUU-------AUCUCAACAUAG-CUUUCCUAG ......((((((((((((.......)))))..((..(((.----------...))).------.))..........-........-------...........)-))))))... ( -22.00) >consensus UCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUGUGG_UG_AUUCUUUUU______UC_AA___AU_AU_UUAAAAAU_CAAUGUAAUACAAAAUAUAAUUUCUAUG ..((((((.....))))))....(((((.....)))))............................................................................ (-16.30 = -16.30 + 0.00)

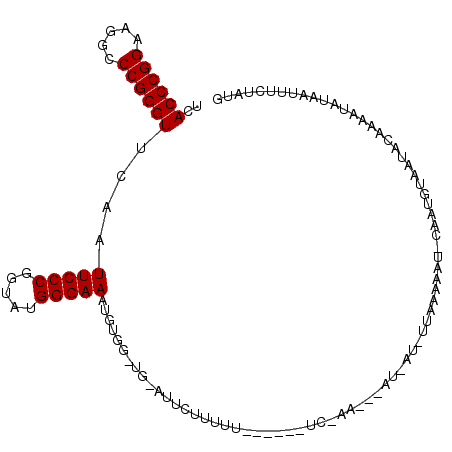
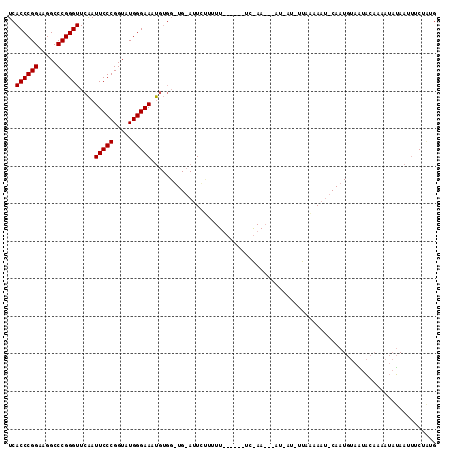
| Location | 1,381,971 – 1,382,082 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.94 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -17.68 |
| Energy contribution | -17.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 4.63 |
| SVM RNA-class probability | 0.999930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381971 111 - 23771897 CAUAGAGAUUAUAUUUUGUACAACAUUGAAUUUUUAAAAUAAU---UUCGAAAAAGAAAAAACAAUUCAACCACAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGA ..((((((((......((.....))...))))))))......(---(((......))))..............((.(((((.....))))).)).((((((.....)))))).. ( -23.10) >DroEre_CAF1 38497 89 - 1 CAGAGAAAUUAUAUUUUGUAUUACAUUGUAUUACA-------------------------AGGAAUGCACCCACAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGA ................((((((...(((.....))-------------------------)..))))))....((.(((((.....))))).)).((((((.....)))))).. ( -24.50) >DroAna_CAF1 38724 89 - 1 CUAGGAAAG-CUAUGUUGAGAU-------AAUUGCAA-AUUAUUUUUUUGA------AAAAAGA----------GUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGA ...((((.(-(((((..(((((-------..((.(((-(.......)))).------...))..----------))))).))))(((((.......))))))).))))...... ( -21.50) >consensus CAAAGAAAUUAUAUUUUGUAAUACAUUG_AUUUCAAA_AU_AU___UU_GA______AAAAAGAAU_CA_CCACAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGA .........................................................................((.(((((.....))))).)).((((((.....)))))).. (-17.68 = -17.57 + -0.11)

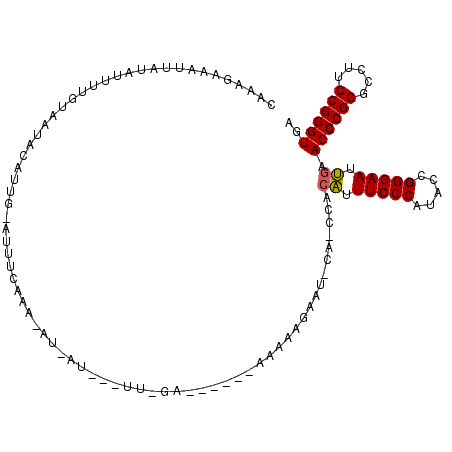
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:26 2006