
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,670,081 – 13,670,313 |
| Length | 232 |
| Max. P | 0.997917 |

| Location | 13,670,081 – 13,670,195 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.08 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -33.13 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13670081 114 - 23771897 ---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGUUCGUGUCGGGUUCCCUGCCACUGGCAGUUGCUCCU---CCU ---....((((((((((((((...(((...)))...)))))))))((((....))))...)))))..((((((....((..(((.((((...)))).)))..)).))))))...---... ( -36.10) >DroSec_CAF1 77471 114 - 1 ---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCUCGUGUCGGGUUCCCUGCUACUGGCAGUUGCUCCU---CCU ---(((.((((((((((((((...(((...)))...)))))))))((((....))))...)))))..))).....((((...((((((...........))))))...))))..---... ( -37.20) >DroSim_CAF1 87613 117 - 1 ---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCUCGUGUCGGGUUCCCUGCCACUGGCAGUUGCUCCUCCUCCU ---....((((((((((((((...(((...)))...)))))))))((((....))))...)))))..((((((....(((.(((.((((...)))).))).))).))))))......... ( -38.30) >DroEre_CAF1 82673 117 - 1 ---GCUCCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCUCGUGUCGGGUUCCCAGCCAGUGGCAGUUGCACCUCCCCCC ---((..((((.((((((.....(((((.(((....)))(((..(((((....)))))..))).)))))......(((((((...))))..))).).)))))))))..)).......... ( -38.70) >DroYak_CAF1 86022 108 - 1 GCUGCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCUCCU------------GCCAGUGGCAGUUGCUCCUCUCCCC ((((...((((((((((((((...(((...)))...)))))))))((((....))))...))))).)))).....((((..((------------(((...)))))..))))........ ( -41.30) >consensus ___GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCUCGUGUCGGGUUCCCUGCCACUGGCAGUUGCUCCUC__CCU .......((((((((((((((...(((...)))...)))))))))((((....))))...)))))...(((((....(((.(((.((((...)))).))).))).))))).......... (-33.13 = -33.56 + 0.43)


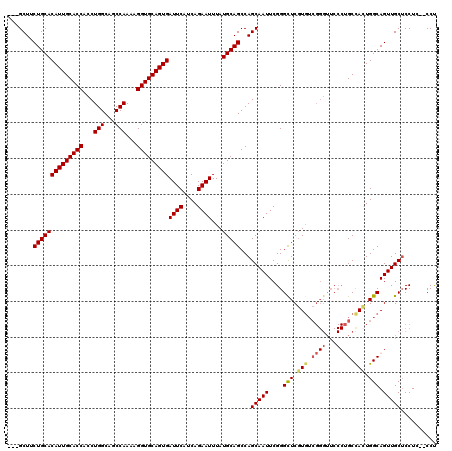
| Location | 13,670,118 – 13,670,235 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13670118 117 + 23771897 AACCCGAAUUGCUGGCUGCAUAAAUUCUGAUGAAUCACUGCACCUUUUGGCUGCCAGGUGGUGCAAUGUGCAGAAGC---AGCAUCCGCCCAAUAAUGUUUCCACGUAAUGCGGCAUCUA .........(((((.(((((((.((((....))))...((((((.(((((...))))).)))))).)))))))...)---))))...(((((...((((....))))..)).)))..... ( -36.50) >DroSec_CAF1 77508 117 + 1 AGCCCGAAUUGCUGGCUGCAUAAAUUCUGAUGAAUCACUGCACCUUUUGGCUGCCAGGUGGUGCAAUGUGCAGAAGC---AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUA .........(((((.(((((((.((((....))))...((((((.(((((...))))).)))))).)))))))...)---))))...(((((...((((....))))..)).)))..... ( -36.50) >DroSim_CAF1 87653 117 + 1 AGCCCGAAUUGCUGGCUGCAUAAAUUCUGAUGAAUCACUGCACCUUUUGGCUGCCAGGUGGUGCAAUGUGCAGAAGC---AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUA .........(((((.(((((((.((((....))))...((((((.(((((...))))).)))))).)))))))...)---))))...(((((...((((....))))..)).)))..... ( -36.50) >DroEre_CAF1 82713 117 + 1 AGCCCGAAUUGCUGGCUGCAUAAAUUCUGAUGAAUCACUGCACCUUUUGGCUGCCAGGUGGUGCAAUGUGCAGGAGC---AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUA .(((...(((((((((((((((.((((....))))...((((((.(((((...))))).)))))).)))))))((((---(((....)).......)))))))).)))))..)))..... ( -36.01) >DroYak_CAF1 86050 120 + 1 AGCCCGAAUUGCUGGCUGCAUAAAUUCUGAUGAAUCACUGCACCUUUUGGCUGCCAGGUGGUGCAAUGUGCAGAAGCAGCAGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUA .(((.....((((..(((((((.((((....))))...((((((.(((((...))))).)))))).))))))).))))(((((....))......((((....))))..))))))..... ( -37.70) >consensus AGCCCGAAUUGCUGGCUGCAUAAAUUCUGAUGAAUCACUGCACCUUUUGGCUGCCAGGUGGUGCAAUGUGCAGAAGC___AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUA .(((...((((((((..((((...(((((......((.((((((.(((((...))))).)))))).))..)))))......((....))......))))..))).)))))..)))..... (-34.20 = -34.24 + 0.04)



| Location | 13,670,118 – 13,670,235 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -37.48 |
| Energy contribution | -37.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13670118 117 - 23771897 UAGAUGCCGCAUUACGUGGAAACAUUAUUGGGCGGAUGCU---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGUU .....(((.((.((.(((....))))).)))))(((((((---(...((((((((((((((...(((...)))...)))))))))((((....))))...))))).)))).))))..... ( -40.80) >DroSec_CAF1 77508 117 - 1 UAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCU .....(((((.....(((....)))......))(((((((---(...((((((((((((((...(((...)))...)))))))))((((....))))...))))).)))).)))).))). ( -41.60) >DroSim_CAF1 87653 117 - 1 UAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCU .....(((((.....(((....)))......))(((((((---(...((((((((((((((...(((...)))...)))))))))((((....))))...))))).)))).)))).))). ( -41.60) >DroEre_CAF1 82713 117 - 1 UAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU---GCUCCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCU .....(((((.....(((....)))......))(((((((---(...((((((((((((((...(((...)))...)))))))))((((....))))...))))).)))).)))).))). ( -41.60) >DroYak_CAF1 86050 120 - 1 UAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCUGCUGCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCU .....(((((.....(((....)))......))(((((((((((((((((..(((((((((...(((...)))...)))))))))......))))).....)))).)))).)))).))). ( -42.90) >consensus UAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU___GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUGCAGUGAUUCAUCAGAAUUUAUGCAGCCAGCAAUUCGGGCU .((...((((.....(((....)))......))))...))...(((.((((((((((((((...(((...)))...)))))))))((((....))))...)))))..))).......... (-37.48 = -37.48 + 0.00)

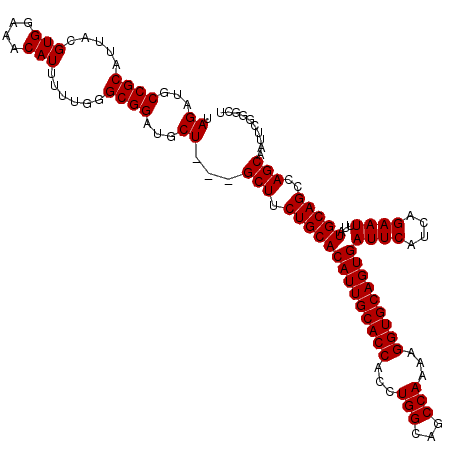

| Location | 13,670,158 – 13,670,273 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -36.24 |
| Consensus MFE | -34.94 |
| Energy contribution | -34.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13670158 115 - 23771897 GUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUAUUGGGCGGAUGCU---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUG .....((((((((((((.(((....((((........)))).))).)).....(((....)))....))))))))))(---(((..(((.....)))......))))(((....))). ( -33.70) >DroSec_CAF1 77548 115 - 1 GUUCCGCGUUCGCCCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUG .....((((((((((((.(((....((((........)))).))).)).....(((....)))....))))))))))(---(((..(((.....)))......))))(((....))). ( -36.70) >DroSim_CAF1 87693 115 - 1 GUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU---GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUG .....((((((((((((.(((....((((........)))).))).)).....(((....)))....))))))))))(---(((..(((.....)))......))))(((....))). ( -33.70) >DroEre_CAF1 82753 115 - 1 GCUCCGCGUUCGCCCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU---GCUCCUGCACAUUGCACCACCUGGCAGCCAAAAGGUG ((...((((((((((((.(((....((((........)))).))).)).....(((....)))....))))))))))(---((....)))....))..(((((..........))))) ( -38.40) >DroYak_CAF1 86090 118 - 1 GUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCUGCUGCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUG ...((((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))).((((((..(((.....)))......)))))).....)).. ( -38.70) >consensus GUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU___GCUUCUGCACAUUGCACCACCUGGCAGCCAAAAGGUG .....((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))).........(((.....))).(((((..........))))) (-34.94 = -34.70 + -0.24)


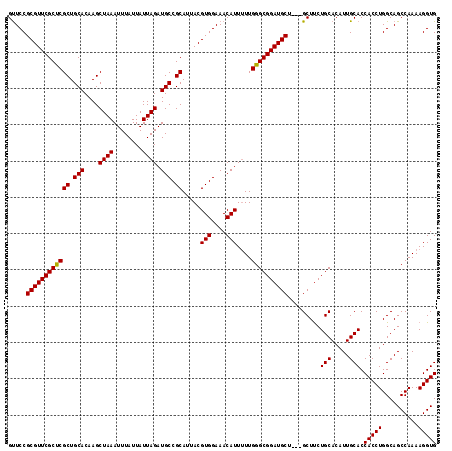
| Location | 13,670,195 – 13,670,313 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -22.72 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13670195 118 + 23771897 AGCAUCCGCCCAAUAAUGUUUCCACGUAAUGCGGCAUCUAAUAAUAAAUUUAGCUUGUGCAGCGAGCGAACGCGGAACCAAAUUUCGCAGAAUGCAUAAUUCAAAAUAAUUCAAAACA .((((..(((((...((((....))))..)).))).(((.............(((((.....)))))....((((((.....)))))))))))))....................... ( -23.60) >DroSec_CAF1 77585 118 + 1 AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUAAUAAUAAAUUUAGCUUGUGCAGCGGGCGAACGCGGAACCAAAUUUCGCAGAAUGCAUAAUUCAAAAUAAUUCAAAACA .((((.(((((....((((....))))...((.((((((((........))))...)))).)))))))...((((((.....))))))...))))....................... ( -28.70) >DroSim_CAF1 87730 118 + 1 AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUAAUAAUAAAUUUAGCUUGUGCAGCGAGCGAACGCGGAACCAAAUUUCGCAGAAUGCAUAAUUCAAAAUAAUUCAAAACA .((((..(((((...((((....))))..)).))).(((.............(((((.....)))))....((((((.....)))))))))))))....................... ( -23.60) >DroEre_CAF1 82790 118 + 1 AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUAAUAAUAAAUUUAGCUUGUGCAGCGGGCGAACGCGGAGCCAAAAUUCGCAGAAUGCAUAAUUCAAAAUAAUUCAAAACA .((((.(((((....((((....))))...((.((((((((........))))...)))).)))))))...(((((.......)))))...))))....................... ( -28.50) >DroYak_CAF1 86130 118 + 1 AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUAAUAAUAAAUUUAGCUUGUGCAGCGAGCGAACGCGGAACCAGAAUUCGCAGAAUGCAUAAUUCAAAAUAAUUCAAAACA .((((..(((((...((((....))))..)).))).(((.............(((((.....)))))....(((((.......))))))))))))....................... ( -22.60) >consensus AGCAUCCGCCCAAAAAUGUUUCCACGUAAUGCGGCAUCUAAUAAUAAAUUUAGCUUGUGCAGCGAGCGAACGCGGAACCAAAUUUCGCAGAAUGCAUAAUUCAAAAUAAUUCAAAACA .((((..(((((...((((....))))..)).))).(((.............(((((.....)))))....((((((.....)))))))))))))....................... (-22.72 = -22.88 + 0.16)



| Location | 13,670,195 – 13,670,313 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -30.78 |
| Energy contribution | -31.14 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13670195 118 - 23771897 UGUUUUGAAUUAUUUUGAAUUAUGCAUUCUGCGAAAUUUGGUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUAUUGGGCGGAUGCU ..............(..((((.(((.....))).))))..)....((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))). ( -32.20) >DroSec_CAF1 77585 118 - 1 UGUUUUGAAUUAUUUUGAAUUAUGCAUUCUGCGAAAUUUGGUUCCGCGUUCGCCCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU ..............(..((((.(((.....))).))))..)....((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))). ( -35.20) >DroSim_CAF1 87730 118 - 1 UGUUUUGAAUUAUUUUGAAUUAUGCAUUCUGCGAAAUUUGGUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU ..............(..((((.(((.....))).))))..)....((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))). ( -32.20) >DroEre_CAF1 82790 118 - 1 UGUUUUGAAUUAUUUUGAAUUAUGCAUUCUGCGAAUUUUGGCUCCGCGUUCGCCCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU .(((..(((((.....((((.....))))....))))).)))...((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))). ( -32.60) >DroYak_CAF1 86130 118 - 1 UGUUUUGAAUUAUUUUGAAUUAUGCAUUCUGCGAAUUCUGGUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU ......((((((....(((((.(((.....)))))))))))))).((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))). ( -33.50) >consensus UGUUUUGAAUUAUUUUGAAUUAUGCAUUCUGCGAAAUUUGGUUCCGCGUUCGCUCGCUGCACAAGCUAAAUUUAUUAUUAGAUGCCGCAUUACGUGGAAACAUUUUUGGGCGGAUGCU ..............(..((((.(((.....))).))))..)....((((((((((((.(((....((((........)))).))).)).....(((....)))....)))))))))). (-30.78 = -31.14 + 0.36)

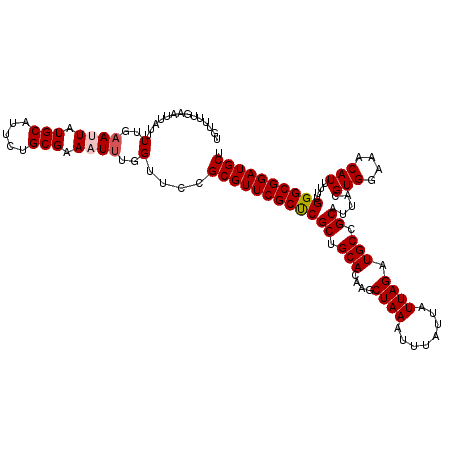
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:05 2006