
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,669,679 – 13,669,858 |
| Length | 179 |
| Max. P | 0.942646 |

| Location | 13,669,679 – 13,669,799 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13669679 120 - 23771897 UUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUUGCCCGGGCUCGUAAACACCUCCGAAAGUCCUCAACCGCCA ..............((((..((((((((......))))))))..)))).((((.(..(((((((((((((((...))))))).((((...(....)...))))..))).)))))))))). ( -34.50) >DroSec_CAF1 77069 120 - 1 UUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUUGCCCGGGCUCGUAAACACCUCCGAAAGUCCUCAACCACCA ............(.((((..((((((((......))))))))..)))).)(.(((..(((((((((((((((...))))))).((((...(....)...))))..))).))))).)))). ( -30.20) >DroSim_CAF1 87211 120 - 1 UUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUUGCCCGGGCUCGUAAACACCUCCGAAAGUCCUCAACCACAA ............(.((((..((((((((......))))))))..)))).)..(((..(((((((((((((((...))))))).((((...(....)...))))..))).))))).))).. ( -31.00) >DroEre_CAF1 82273 120 - 1 UUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUUGCCUGGGCUCGUAAACACCUCCGAAAGUCCUCAACCACCG .....(((......((((..((((((((......))))))))..)))).((.((....((((.(((((((((...)))))))..)).))))...)).)).......)))........... ( -30.00) >DroYak_CAF1 85583 120 - 1 UUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUUGCCUGGGCUCGUAAACACCUCCGAAAGUCCUCGACCACCA ..............((((..((((((((......))))))))..))))(((((.((((((((.(((((((((...)))))))..)).)))(....)........)))))..)).)))... ( -30.90) >consensus UUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUUGCCCGGGCUCGUAAACACCUCCGAAAGUCCUCAACCACCA .....(((......((((..((((((((......))))))))..)))).((.((....((((.(((((((((...)))))))..)).))))...)).)).......)))........... (-30.00 = -30.00 + 0.00)


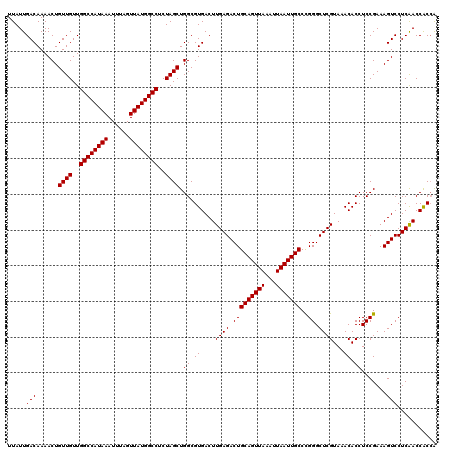
| Location | 13,669,719 – 13,669,824 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.51 |
| Mean single sequence MFE | -19.19 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13669719 105 + 23771897 AAUUAAUUUAACUGCAGUCUCAAGUCACGCCAGCUAGAGGCCAUAACUAAAUUUAUGGCCAACAACAGUUUUGUCAAUAACUUUUAUGCGCCUUCCU---------CCUGCUCC------ .............((((....(((...(((........((((((((......))))))))......((((........)))).....))).)))...---------.))))...------ ( -20.50) >DroSec_CAF1 77109 105 + 1 AAUUAAUUUAACUGCAGUCUCAAGUCACGCCAGCUAGAGGCCAUAACUAAAUUUAUGGCCAACAACAGUUUUGUCAAUAACUUUUAUGCGCCAUCCU---------CCAGCUCC------ .(((.((..(((((..((.((.(((.......))).))((((((((......)))))))).))..)))))..)).))).........((........---------...))...------ ( -17.70) >DroSim_CAF1 87251 105 + 1 AAUUAAUUUAACUGCAGUCUCAAGUCACGCCAGCUAGAGGCCAUAACUAAAUUUAUGGCCAACAACAGUUUUGUCAAUAACUUUUAUGCGCCAUCCU---------CCUGCUCC------ .............((((..........(((........((((((((......))))))))......((((........)))).....))).......---------.))))...------ ( -19.07) >DroEre_CAF1 82313 105 + 1 AAUUAAUUUAACUGCAGUCUCAAGUCACGCCAGCUAGAGGCCAUAACUAAAUUUAUGGCCAACAACAGUUUUGUCAAUAACUUUUAUGCGCCAUCCU---------CCUGCUCC------ .............((((..........(((........((((((((......))))))))......((((........)))).....))).......---------.))))...------ ( -19.07) >DroYak_CAF1 85623 120 + 1 AAUUAAUUUAACUGCAGUCUCAAGUCACGCCAGCUAGAGGCCAUAACUAAAUUUAUGGCCAACAACAGUUUUGUCAAUAACUUUUAUGCGCCAUCCCCCUGAUCCUCCUGCUCCUGCUCC .............((((......(((((((........((((((((......))))))))......((((........)))).....))).........))))....))))......... ( -19.60) >consensus AAUUAAUUUAACUGCAGUCUCAAGUCACGCCAGCUAGAGGCCAUAACUAAAUUUAUGGCCAACAACAGUUUUGUCAAUAACUUUUAUGCGCCAUCCU_________CCUGCUCC______ .(((.((..(((((..((.((.(((.......))).))((((((((......)))))))).))..)))))..)).))).......................................... (-17.50 = -17.50 + 0.00)


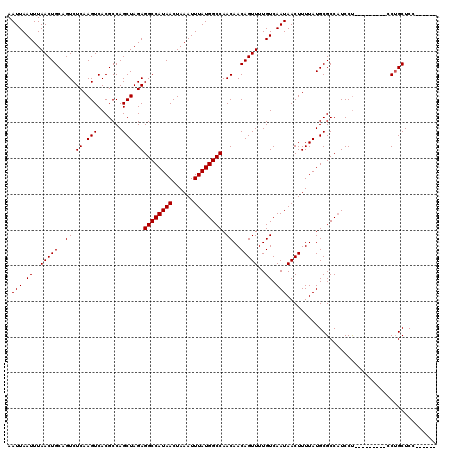
| Location | 13,669,719 – 13,669,824 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.51 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13669719 105 - 23771897 ------GGAGCAGG---------AGGAAGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUU ------...((((.---------...(((((((.....(((((..(((((.....)))))((((((((......))))))))..))))).))))..)))....))))............. ( -28.30) >DroSec_CAF1 77109 105 - 1 ------GGAGCUGG---------AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUU ------(.((((.(---------((....((((.....(((((..(((((.....)))))((((((((......))))))))..))))).))))..))).)).)).)............. ( -28.70) >DroSim_CAF1 87251 105 - 1 ------GGAGCAGG---------AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUU ------...((((.---------(((.(.((((.....(((((..(((((.....)))))((((((((......))))))))..))))).)))).))))....))))............. ( -29.10) >DroEre_CAF1 82313 105 - 1 ------GGAGCAGG---------AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUU ------...((((.---------(((.(.((((.....(((((..(((((.....)))))((((((((......))))))))..))))).)))).))))....))))............. ( -29.10) >DroYak_CAF1 85623 120 - 1 GGAGCAGGAGCAGGAGGAUCAGGGGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUU ...((((...((((...(((.....))).((((.....(((((..(((((.....)))))((((((((......))))))))..))))).))))..))))...))))............. ( -33.00) >consensus ______GGAGCAGG_________AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACUUGAGACUGCAGUUAAAUUAAUU .........((((..........(((.(.((((.....(((((..(((((.....)))))((((((((......))))))))..))))).)))).))))....))))............. (-25.14 = -25.38 + 0.24)


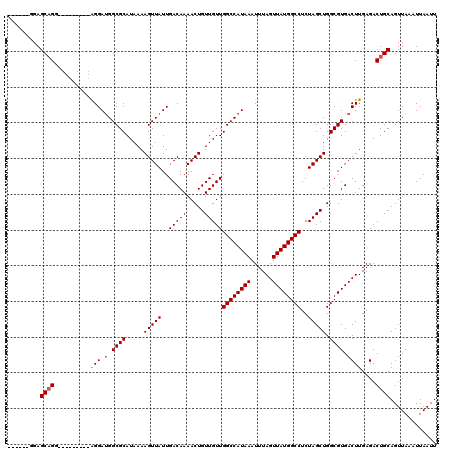
| Location | 13,669,759 – 13,669,858 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.13 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.48 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13669759 99 - 23771897 GAUUAAUGAAGUGCCGGCUAGAUUAGGUGGCACA------------GGAGCAGG---------AGGAAGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGG .............(((((((((((..(((((.((------------(.(((((.---------.(..((..((......))..))..)....))))).)))))))).))))))))..))) ( -23.60) >DroSec_CAF1 77149 99 - 1 GAUUAAUGAAGUGCCGGCUAGAUUAGGUGGCCCA------------GGAGCUGG---------AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGG ............((((.(((...))).))))(((------------..((((((---------(..(((((.(((((.((((........)))).))).)))))))...))))))).))) ( -26.80) >DroSim_CAF1 87291 99 - 1 GAUUAAUGAAGUGCCGGCUAGAUUAGGUGGCCCA------------GGAGCAGG---------AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGG .............(((((((((((..((((((.(------------(.(((((.---------..((((((........)))))).......))))).)))))))).))))))))..))) ( -26.80) >DroEre_CAF1 82353 99 - 1 GAUUAAUGAAGUGCCGGCUAGAUUGGGUGGCACA------------GGAGCAGG---------AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGG .............(((((((((((..(((((.((------------(.(((((.---------..((((((........)))))).......))))).)))))))).))))))))..))) ( -28.00) >DroYak_CAF1 85663 120 - 1 GAUUAAUGAAGUGCCGGCUAGAUUAGGUGGCACAUGGCCAGGAGCAGGAGCAGGAGGAUCAGGGGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGG ((((((....((((((.(((...))).))))))((((((((.(((((((.........))..(..((((((........))))))..)....))))).))))))))....)))))).... ( -39.20) >consensus GAUUAAUGAAGUGCCGGCUAGAUUAGGUGGCACA____________GGAGCAGG_________AGGAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGG ((((((....((((((.(((...))).)))))).................................(((((.(((((.((((........)))).))).)))))))....)))))).... (-21.04 = -21.48 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:59 2006