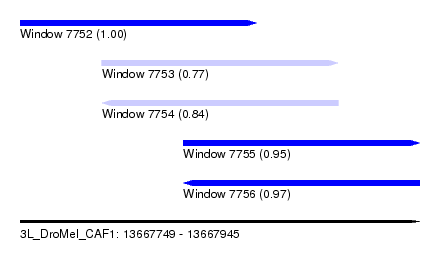
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,667,749 – 13,667,945 |
| Length | 196 |
| Max. P | 0.995534 |

| Location | 13,667,749 – 13,667,865 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.22 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13667749 116 + 23771897 AAGGAAAGUUGACGCGGUGGGGCCCAGACAAUGUUGGCUGUCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCUGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAA .......(((..(((..(((....((((..((((..(((((((......)).........)))))..))))))))....)))..))).)))..(----((((((....)))))))..... ( -37.80) >DroSec_CAF1 75202 116 + 1 AAGGAAAGUUGACGCGGUGGGGCCCAGACAAUGUUGGCUGUCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCAGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAA .((((.(((..(((..((.(....)..))..)))..))).))))((((((.(((((......)))))...(((((....))))).))))))..(----((((((....)))))))..... ( -39.60) >DroSim_CAF1 85472 116 + 1 AAGGAAAGUUGACGCGGUGGGGCCCAGACAAUGUUGGCUGUCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCAGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAA .((((.(((..(((..((.(....)..))..)))..))).))))((((((.(((((......)))))...(((((....))))).))))))..(----((((((....)))))))..... ( -39.60) >DroEre_CAF1 80315 120 + 1 AAGGAAAGUUGACGCGGUGGGGCCCAGACAAUGUUGGCUGUCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCUGCAAACUGAGGCGGGACGAGGGAGGGCGGCAGCAGCUGCCUAUAAA ......(((..(((..((.(....)..))..)))..)))(((((((((..((((((......)))))............)..)))))))))......(((((((....)))))))..... ( -38.30) >DroYak_CAF1 83608 116 + 1 AAGGAAAGUUGACGCGCUGGGGCCCAGACAAUGUUGGCUGUCCUGUCCUGGCAAUUAAAACCAGUUGACAUUCUGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAA .((((.(((..(((..((((...))))....)))..))).))))(((((((.........))))..))).((((((........))))))...(----((((((....)))))))..... ( -41.30) >consensus AAGGAAAGUUGACGCGGUGGGGCCCAGACAAUGUUGGCUGUCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCUGCAAACUGAAGCGGGACGAA____GGCAGCAGCAGCUGCCUAUAAA .((((.(((..(((..((.(....)..))..)))..))).))))((((((.(((((......)))))...(((........))).)))))).......((((((....))))))...... (-34.50 = -34.22 + -0.28)



| Location | 13,667,789 – 13,667,905 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.43 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -27.86 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13667789 116 + 23771897 UCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCUGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUG ....(.(((((((.........((((..(.((((((........)))))).).(----((((((....)))))))...))))..(((((....).)))).........))))))).)... ( -34.80) >DroSec_CAF1 75242 116 + 1 UCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCAGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUG ....(.(((((((..........(((....(((((....)))))....)))..(----((((((....))))))).........(((((....).)))).........))))))).)... ( -32.50) >DroSim_CAF1 85512 116 + 1 UCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCAGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUG ....(.(((((((..........(((....(((((....)))))....)))..(----((((((....))))))).........(((((....).)))).........))))))).)... ( -32.50) >DroEre_CAF1 80355 120 + 1 UCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCUGCAAACUGAGGCGGGACGAGGGAGGGCGGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUG .(((.(((((.(((((......)))))...((((((........))))))))))).)))((((((((((((............)))(((....)))............))))..))))). ( -34.90) >DroYak_CAF1 83648 116 + 1 UCCUGUCCUGGCAAUUAAAACCAGUUGACAUUCUGCAAACUGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUG .........(((..........((((..(.((((((........)))))).).(----((((((....)))))))...))))..(((((....).))))...............)))... ( -31.60) >consensus UCCUGUCUUGGCAAUUAAAAACAGUUGACAUUCUGCAAACUGAAGCGGGACGAA____GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUG ....(.(((((((........((((((.(.((((((........))))))........((((((....))))))..........))))).))(((((.....))))).))))))).)... (-27.86 = -28.30 + 0.44)

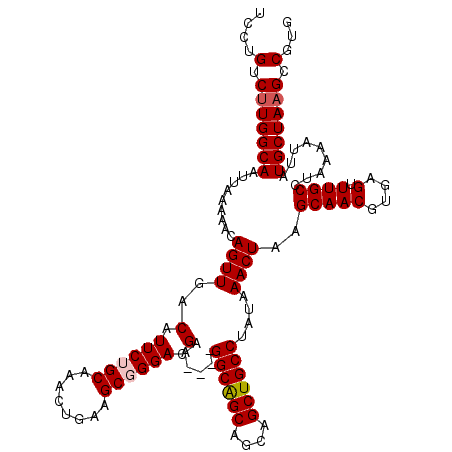
| Location | 13,667,789 – 13,667,905 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.43 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13667789 116 - 23771897 CACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCAGUUUGCAGAAUGUCAACUGUUUUUAAUUGCCAAGACAGGA ...(((..(((((((..((((((((.......)))))))).))))(((((((....))))))----).......))).(((((.((.....)).))))).........)))......... ( -32.10) >DroSec_CAF1 75242 116 - 1 CACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCAGUUUGCUGAAUGUCAACUGUUUUUAAUUGCCAAGACAGGA ...(((...((((((..((((((((.......)))))))).))))(((((((....))))))----).......))(((((....))))).)))..(((((((........))))))).. ( -33.70) >DroSim_CAF1 85512 116 - 1 CACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCAGUUUGCUGAAUGUCAACUGUUUUUAAUUGCCAAGACAGGA ...(((...((((((..((((((((.......)))))))).))))(((((((....))))))----).......))(((((....))))).)))..(((((((........))))))).. ( -33.70) >DroEre_CAF1 80355 120 - 1 CACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCCGCCCUCCCUCGUCCCGCCUCAGUUUGCAGAAUGUCAACUGUUUUUAAUUGCCAAGACAGGA ...(((.....((((..((((((((.......)))))))).)))).(((((...))))))))...(((.(((..((..(((((.((.....)).))))).........))...)))))). ( -28.50) >DroYak_CAF1 83648 116 - 1 CACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCAGUUUGCAGAAUGUCAACUGGUUUUAAUUGCCAGGACAGGA ...(((.....((((..((((((((.......)))))))).))))(((((((....))))))----)..)))((((........))....((((..(((((.......))))))))))). ( -34.30) >consensus CACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC____UUCGUCCCGCUUCAGUUUGCAGAAUGUCAACUGUUUUUAAUUGCCAAGACAGGA ...(((...((((((..((((((((.......)))))))).)))).((((((....))))))............))..(((((.((.....)).))))).........)))......... (-29.16 = -29.36 + 0.20)

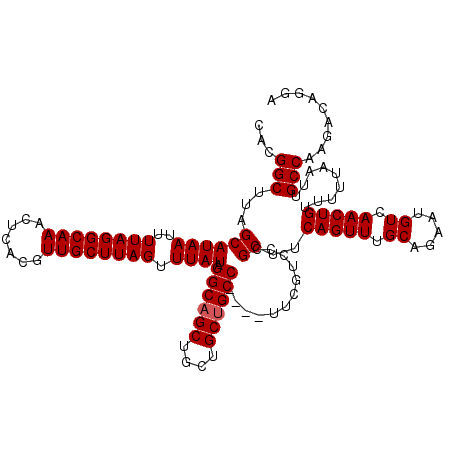
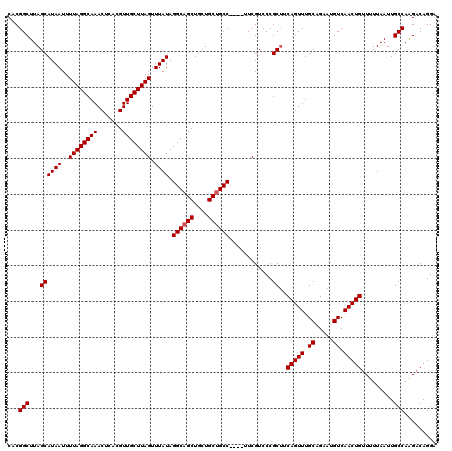
| Location | 13,667,829 – 13,667,945 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13667829 116 + 23771897 UGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUGCUAAGAAAAGCGCAAAAGCACUGCGAGAAAUUAAUAUCAA (((.((((.....(----((((((....))))))).........(((((....).)))).........((((.....(((((......)))))...))))))))((....))....))). ( -32.30) >DroSec_CAF1 75282 116 + 1 UGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUGCUAAGAAAAGCGCAAAAGCACUGAGAGAAAUUUAUAUCAA (((.((((.....(----((((((....))))))).........(((((....).))))................))(((((......)))))....))....(((....)))...))). ( -30.30) >DroSim_CAF1 85552 116 + 1 UGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUGCUAAGAAAAGCGCAAAAGCAUUGAGAGAAAUUUAUAUCAA (((.((((.....(----((((((....))))))).........(((((....).))))................))(((((......)))))....))....(((....)))...))). ( -30.30) >DroEre_CAF1 80395 119 + 1 UGAGGCGGGACGAGGGAGGGCGGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUGCUAAGAAAAGAGCAA-UGCACGGCGAGAAAUAAAUAUGAA ..(((((..........(((((((....)))))))...........(((....))))))))....((((.....(((((((..............-.))))))).....))))....... ( -36.16) >DroYak_CAF1 83688 112 + 1 UGAAGCGGGACGAA----GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUGCUAAGAAAAGAGCAA-AGCACUGCGAGAAAUAAAUAU--- ....((((.....(----((((((....))))))).........(((((....).))))...............((..((((........)))).-.)).)))).............--- ( -31.00) >consensus UGAAGCGGGACGAA____GGCAGCAGCAGCUGCCUAUAAACUAAGCAACGUGAGUUUGCCUAAAAUUAUGCUAAGCCGUGCUAAGAAAAGCGCAAAAGCACUGCGAGAAAUUAAUAUCAA ....((((..........((((((....))))))......((.((((.....(((((.....))))).)))).))))(((((......)))))....))..................... (-24.52 = -24.76 + 0.24)


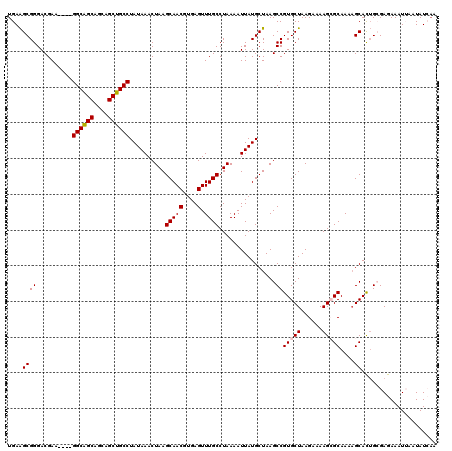
| Location | 13,667,829 – 13,667,945 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13667829 116 - 23771897 UUGAUAUUAAUUUCUCGCAGUGCUUUUGCGCUUUUCUUAGCACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCA ................((.(((((..............))))).))..(((((((..((((((((.......)))))))).))))(((((((....))))))----).......)))... ( -30.04) >DroSec_CAF1 75282 116 - 1 UUGAUAUAAAUUUCUCUCAGUGCUUUUGCGCUUUUCUUAGCACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCA ....(((((((........(((((.(((.(((......))).)))...))))).....(((((((.......))))))))))))))((((((....))))))----.............. ( -29.50) >DroSim_CAF1 85552 116 - 1 UUGAUAUAAAUUUCUCUCAAUGCUUUUGCGCUUUUCUUAGCACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCA ....(((((((........(((((.(((.(((......))).)))...))))).....(((((((.......))))))))))))))((((((....))))))----.............. ( -29.20) >DroEre_CAF1 80395 119 - 1 UUCAUAUUUAUUUCUCGCCGUGCA-UUGCUCUUUUCUUAGCACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCCGCCCUCCCUCGUCCCGCCUCA ................(((((((.-..............))))))).............((((........((((((((.....)))))))).((....)).............)))).. ( -28.46) >DroYak_CAF1 83688 112 - 1 ---AUAUUUAUUUCUCGCAGUGCU-UUGCUCUUUUCUUAGCACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC----UUCGUCCCGCUUCA ---.............((.(((((-..(.......)..))))).))..(((((((..((((((((.......)))))))).))))(((((((....))))))----).......)))... ( -30.40) >consensus UUGAUAUUAAUUUCUCGCAGUGCUUUUGCGCUUUUCUUAGCACGGCUUAGCAUAAUUUUAGGCAAACUCACGUUGCUUAGUUUAUAGGCAGCUGCUGCUGCC____UUCGUCCCGCUUCA ...................(((((..............)))))(((...((((((..((((((((.......)))))))).)))).((((((....)))))).......))...)))... (-23.44 = -23.88 + 0.44)

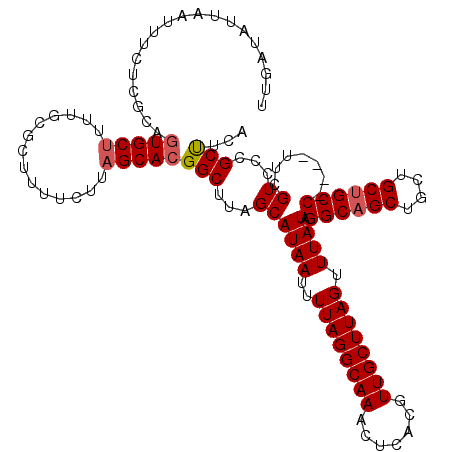

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:49 2006