| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
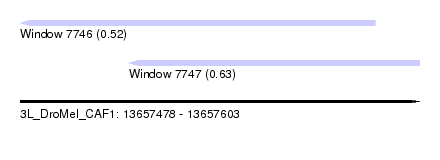
| Location | 13,657,478 – 13,657,603 |
| Length | 125 |
| Max. P | 0.631280 |

| Location | 13,657,478 – 13,657,589 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -19.27 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
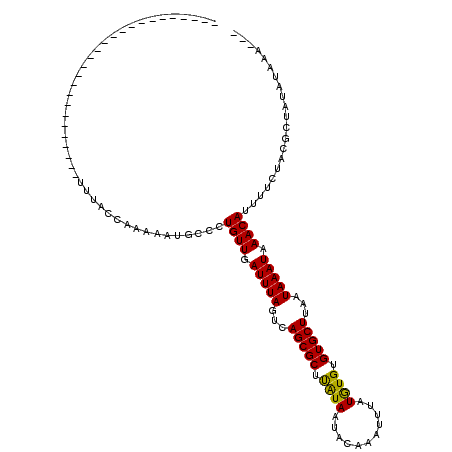
>3L_DroMel_CAF1 13657478 111 - 23771897 CCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA----CUCAAAUACAGCGUCACAACAUCAGUUACAAAAG----- ..((((((((((...(((((.(((((.......)))))...)))))...)))))...........(((((.(((...----.....))).)))))..))))).............----- ( -19.40) >DroPse_CAF1 49919 111 - 1 UUUGUUGAUUUAGUCAGCGCUCAUAAUACAAAUUUAUAUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA----CGCAAAUAUGUUGGAAAAACAACAGCAACGCAAC----- ..((((.(((((...(((((.((((...........)))).)))))...))))).))))..................----.((.....(((((......))))).....))...----- ( -21.20) >DroEre_CAF1 70012 111 - 1 CCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAGUUUCUACGCUAUAUAAA----CUCAAAUACAGCGUCACUACAUCAGUUACAAAAA----- .(((((.(((((...(((((.(((((.......)))))...)))))...))))).))))).....(((((.(((...----.....))).)))))....................----- ( -20.80) >DroYak_CAF1 73058 111 - 1 CCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA----CUCAAAUACGGCGUCACUACAUCAGUUACAAAAG----- .(((.(((((((...(((((.(((((.......)))))...)))))...)))))...........((((..(((...----.....)))..)))).....)).))).........----- ( -18.60) >DroAna_CAF1 108070 110 - 1 AUUGUUUAUUUAACCAGCGCUUGUAAUUCAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCCACGCUACUUAAAUAUACUAAAAUA--ACAUCGCAA--------ACCAAAAACCAU ..((((((((((...(((((.(((((.......)))))...)))))...))))))))))..............................--.........--------............ ( -14.40) >DroPer_CAF1 51146 111 - 1 UUUGUUGAUUUAGUCAGCGCUCAUAAUACAAAUUUAUAUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA----CGCAAAUAUGUUGGAAAAACAACAGCAACGCAAC----- ..((((.(((((...(((((.((((...........)))).)))))...))))).))))..................----.((.....(((((......))))).....))...----- ( -21.20) >consensus CCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA____CUCAAAUACGGCGUCACAACAUCAGUUACAAAAA_____ ..((((.(((((...(((((.((((...........)))).)))))...))))).))))............................................................. (-11.65 = -11.07 + -0.58)

| Location | 13,657,512 – 13,657,603 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.31 |
| Mean single sequence MFE | -18.08 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13657512 91 - 23771897 --------------------------UUUACCAAAAAUGCCCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA--- --------------------------................((((.(((((...(((((.(((((.......)))))...)))))...))))).))))..................--- ( -12.50) >DroPse_CAF1 49953 117 - 1 GGCGAAACCCAACCCUCCAGCUGGGGGCUACUAAAUACGCUUUGUUGAUUUAGUCAGCGCUCAUAAUACAAAUUUAUAUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA--- ((((.........(((((....)))))...............((((.(((((...(((((.((((...........)))).)))))...))))).)))).......)))).......--- ( -25.80) >DroEre_CAF1 70046 91 - 1 --------------------------UUUACCAAAAAUGCCCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAGUUUCUACGCUAUAUAAA--- --------------------------...............(((((.(((((...(((((.(((((.......)))))...)))))...))))).))))).................--- ( -15.30) >DroYak_CAF1 73092 91 - 1 --------------------------UUUACCAAAAAUGCCCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA--- --------------------------................((((.(((((...(((((.(((((.......)))))...)))))...))))).))))..................--- ( -12.50) >DroAna_CAF1 108100 93 - 1 --------------------------UUUACCAAA-AGGCAUUGUUUAUUUAACCAGCGCUUGUAAUUCAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCCACGCUACUUAAAUAU --------------------------.........-.((...((((((((((...(((((.(((((.......)))))...)))))...))))))))))....))............... ( -16.60) >DroPer_CAF1 51180 117 - 1 GGCGAAACCCAACCCUCCAGCUGGGGGCUACUAAAUACGCUUUGUUGAUUUAGUCAGCGCUCAUAAUACAAAUUUAUAUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA--- ((((.........(((((....)))))...............((((.(((((...(((((.((((...........)))).)))))...))))).)))).......)))).......--- ( -25.80) >consensus __________________________UUUACCAAAAAUGCCCUGUUGAUUUAGUCAGCGCUUAUAAUACAAAUUUAUGUGUGUGCUUAAUAAAUAAACAUUUUCUACGCUAUAUAAA___ ..........................................((((.(((((...(((((.((((...........)))).)))))...))))).))))..................... (-11.65 = -11.07 + -0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:41 2006