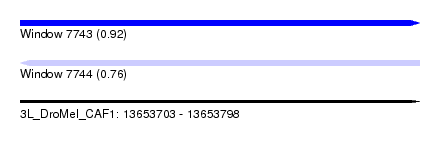
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,653,703 – 13,653,798 |
| Length | 95 |
| Max. P | 0.923286 |

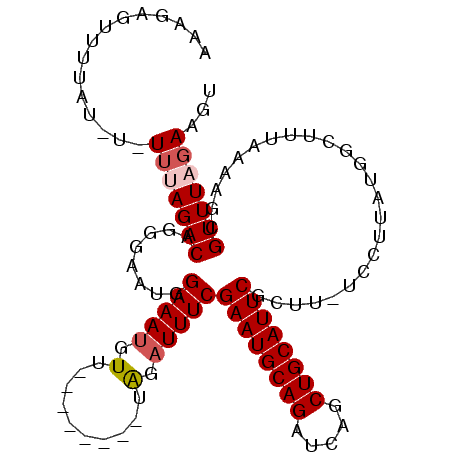
| Location | 13,653,703 – 13,653,798 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.16 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13653703 95 + 23771897 ACUUCUAAACGCUUUUAAAUCCAUAAGGA-AAGCGAAUGCAGCUGAUCUGCAUUCGAAAUCUA--------AACAUUUCCAUUCCCUGUCUAAA-A-AUAAAAUUC--- ..........((((.....(((....)))-))))((((((((.....))))))))(((((...--------...)))))...............-.-.........--- ( -17.10) >DroSec_CAF1 61066 98 + 1 UCUUCUAAACGCUUUUAAAGCCAUAAGGA-AAGCGAAUGCAGCUGAUCUGCAUUCGAAAUCUA--------AACGUUUCCAUUCCCUGUCUAAA-A-AUAAAACUCUCU ..........(((.....))).(((.(((-(...((((((((.....))))))))(((((...--------...)))))..)))).))).....-.-............ ( -17.50) >DroSim_CAF1 71279 100 + 1 ACUUCUAAACACUUUGGAGGCCAUAACGCCCCGCGAAUGCAGCUGAUCUGCAUUCGAAAUCUA--------AACAUUUCCAUUCCCUGUCUAAAAA-AUAAAACUCUCU ..............(((((((......)))...(((((((((.....))))))))).......--------......))))...............-............ ( -19.90) >DroEre_CAF1 63811 99 + 1 ACUUCUAAACGCUUUUAAAGCCGCACGGA-AAGUGAAUGCAGCUGAUCUGCAUUCGAAAUCUU--------AACAUUUCCAUUCCCUGUCUGCA-AAAAAAGAAUCAUU ...........(((((...((.(((.(((-(...((((((((.....))))))))(((((...--------...)))))..)))).)))..)).-..)))))....... ( -21.70) >DroYak_CAF1 68926 106 + 1 ACUUCUAAACGCUUUUAAAGCCGCACGGA-GAGUGAAUGCAGCUGAUCUGCAUUCGAAAUCCUAACAUCCUAACAUUUCCAUUCGCUGUCUGAA-A-AUAAAACUCUUA ..........(((.....)))....((((-.(((((((((((.....))))))))(((((..((......))..))))).....))).))))..-.-............ ( -20.60) >consensus ACUUCUAAACGCUUUUAAAGCCAUAAGGA_AAGCGAAUGCAGCUGAUCUGCAUUCGAAAUCUA________AACAUUUCCAUUCCCUGUCUAAA_A_AUAAAACUCUCU ..........................(((....(((((((((.....)))))))))...)))............................................... (-13.12 = -13.16 + 0.04)

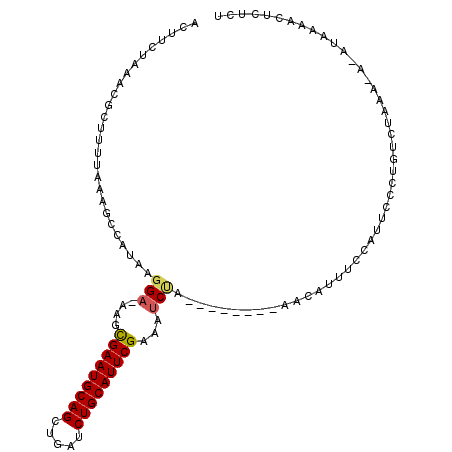
| Location | 13,653,703 – 13,653,798 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.81 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13653703 95 - 23771897 ---GAAUUUUAU-U-UUUAGACAGGGAAUGGAAAUGUU--------UAGAUUUCGAAUGCAGAUCAGCUGCAUUCGCUU-UCCUUAUGGAUUUAAAAGCGUUUAGAAGU ---.......((-(-(((((((........(((((...--------...)))))((((((((.....))))))))((((-(((....))).....)))))))))))))) ( -26.50) >DroSec_CAF1 61066 98 - 1 AGAGAGUUUUAU-U-UUUAGACAGGGAAUGGAAACGUU--------UAGAUUUCGAAUGCAGAUCAGCUGCAUUCGCUU-UCCUUAUGGCUUUAAAAGCGUUUAGAAGA ...........(-(-(((((((((((((((....))))--------)......(((((((((.....)))))))))...-.)))....(((.....)))))))))))). ( -28.90) >DroSim_CAF1 71279 100 - 1 AGAGAGUUUUAU-UUUUUAGACAGGGAAUGGAAAUGUU--------UAGAUUUCGAAUGCAGAUCAGCUGCAUUCGCGGGGCGUUAUGGCCUCCAAAGUGUUUAGAAGU ............-((((((((((.......(((((...--------...)))))((((((((.....))))))))(.(((((......))))))....)))))))))). ( -28.80) >DroEre_CAF1 63811 99 - 1 AAUGAUUCUUUUUU-UGCAGACAGGGAAUGGAAAUGUU--------AAGAUUUCGAAUGCAGAUCAGCUGCAUUCACUU-UCCGUGCGGCUUUAAAAGCGUUUAGAAGU ...(((.(((((..-.((.(.((.((((.((((((...--------...)))))((((((((.....)))))))).).)-))).))).))...))))).)))....... ( -27.40) >DroYak_CAF1 68926 106 - 1 UAAGAGUUUUAU-U-UUCAGACAGCGAAUGGAAAUGUUAGGAUGUUAGGAUUUCGAAUGCAGAUCAGCUGCAUUCACUC-UCCGUGCGGCUUUAAAAGCGUUUAGAAGU ..........((-(-(..((((.(((....(((((.((((....)))).)))))((((((((.....))))))))....-....))).(((.....)))))))..)))) ( -25.00) >consensus AAAGAGUUUUAU_U_UUUAGACAGGGAAUGGAAAUGUU________UAGAUUUCGAAUGCAGAUCAGCUGCAUUCGCUU_UCCUUAUGGCUUUAAAAGCGUUUAGAAGU ...............(((((((........(((((.(..........).)))))((((((((.....))))))))........................)))))))... (-16.04 = -16.68 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:38 2006