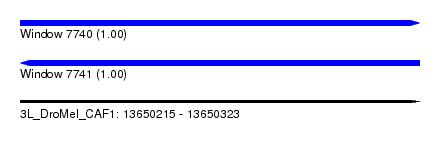
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,650,215 – 13,650,323 |
| Length | 108 |
| Max. P | 0.997538 |

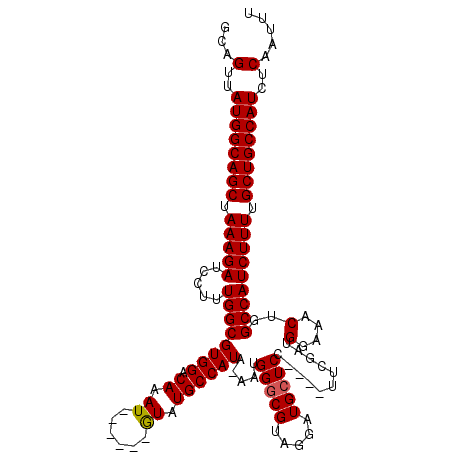
| Location | 13,650,215 – 13,650,323 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.48 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -29.04 |
| Energy contribution | -29.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13650215 108 + 23771897 AAAUUGAGAUGGCAGCAAAAGAUGGCCAGUUUCCAUCGUA----GGAGCAUCCUACGACCAUU-UAUGGCAUAC------AUUUGUCCACGCCAAAGGAUCUUUAGCUGCCAUAACUGC ........((((((((.(((((..((((.......(((((----((.....))))))).....-..))))....------.....(((........)))))))).))))))))...... ( -38.54) >DroSec_CAF1 57608 108 + 1 AAAAUGAGAUGGCAGCAAAAGAUGGCCAGUUUCCAUCGAA----GGAGCAUCCUACGCCCAUU-UAUGGCAUAC------AUUUGUCCACGCCAAAGGAUCUUUAGCUGCCAUAACUGC ........((((((((.(((((..((((........((.(----((.....))).))......-..))))....------.....(((........)))))))).))))))))...... ( -32.09) >DroSim_CAF1 67769 108 + 1 AAAUUGAGAUGGCAGCAAAAGAUGGCCAGUUUCCAUCGAA----GGAGCAUCCUACGCCCAUU-UAUGGCAUAC------AUUUGUCCACGCCAAAGGAUCUUUAGCUGCCAUAACUGC ........((((((((.(((((..((((........((.(----((.....))).))......-..))))....------.....(((........)))))))).))))))))...... ( -32.09) >DroEre_CAF1 60321 109 + 1 AAAUUGAGAUGGCAGCAAAAGAUGGCCAGUUGCCAUCGAA----GGAGCAUCCUACGCCCAUUUUAUGGCAUAU------AUUUGUCCACGCCAAGGGUUCUUUGGCUGCCAUAACUGC ........((((((((.((((((((.(((.((((((.(((----((.((.......)))).))).))))))...------..))).))).(((...)))))))).))))))))...... ( -35.60) >DroYak_CAF1 65551 119 + 1 AAAUUGAGAUGGCAGCAAAAGAUGGCCAGUUUCCAUCGAAGGAAGGAGCAACCUACGCCCAUUUCAUGGCACAUGCUCGAAUUUGUCCACGCCAAAGGUUCUUUGGCUGCCAUAACUGC ........((((((((.(((((((((...(((((......)))))(((((......(((........)))...)))))............)))).....))))).))))))))...... ( -36.40) >consensus AAAUUGAGAUGGCAGCAAAAGAUGGCCAGUUUCCAUCGAA____GGAGCAUCCUACGCCCAUU_UAUGGCAUAC______AUUUGUCCACGCCAAAGGAUCUUUAGCUGCCAUAACUGC ........((((((((....(((((.......))))).......((((.(((((..(((........)))............(((.......)))))))))))).))))))))...... (-29.04 = -29.04 + -0.00)

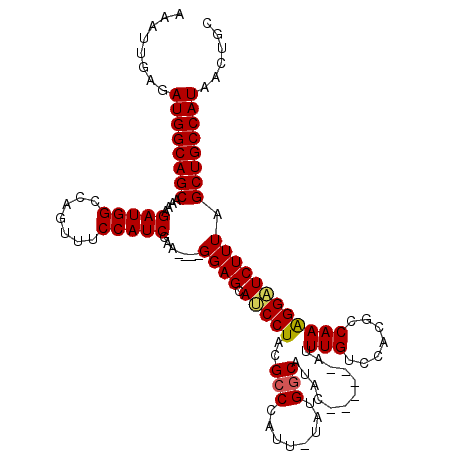
| Location | 13,650,215 – 13,650,323 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.48 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -31.40 |
| Energy contribution | -31.36 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13650215 108 - 23771897 GCAGUUAUGGCAGCUAAAGAUCCUUUGGCGUGGACAAAU------GUAUGCCAUA-AAUGGUCGUAGGAUGCUCC----UACGAUGGAAACUGGCCAUCUUUUGCUGCCAUCUCAAUUU ...(..((((((((.(((((.....(((((((.(....)------.)))))))..-..((((((((((.....))----)))...(....).)))))))))).))))))))..)..... ( -42.70) >DroSec_CAF1 57608 108 - 1 GCAGUUAUGGCAGCUAAAGAUCCUUUGGCGUGGACAAAU------GUAUGCCAUA-AAUGGGCGUAGGAUGCUCC----UUCGAUGGAAACUGGCCAUCUUUUGCUGCCAUCUCAUUUU ...(..((((((((.((((((....(((((((.(....)------.)))))))..-....((((.(((.....))----).)...(....)..))))))))).))))))))..)..... ( -37.30) >DroSim_CAF1 67769 108 - 1 GCAGUUAUGGCAGCUAAAGAUCCUUUGGCGUGGACAAAU------GUAUGCCAUA-AAUGGGCGUAGGAUGCUCC----UUCGAUGGAAACUGGCCAUCUUUUGCUGCCAUCUCAAUUU ...(..((((((((.((((((....(((((((.(....)------.)))))))..-....((((.(((.....))----).)...(....)..))))))))).))))))))..)..... ( -37.30) >DroEre_CAF1 60321 109 - 1 GCAGUUAUGGCAGCCAAAGAACCCUUGGCGUGGACAAAU------AUAUGCCAUAAAAUGGGCGUAGGAUGCUCC----UUCGAUGGCAACUGGCCAUCUUUUGCUGCCAUCUCAAUUU ...(..((((((((.(((((.(((.(((((((.......------.)))))))......)))((.(((.....))----).)).((((.....))))))))).))))))))..)..... ( -40.30) >DroYak_CAF1 65551 119 - 1 GCAGUUAUGGCAGCCAAAGAACCUUUGGCGUGGACAAAUUCGAGCAUGUGCCAUGAAAUGGGCGUAGGUUGCUCCUUCCUUCGAUGGAAACUGGCCAUCUUUUGCUGCCAUCUCAAUUU ...(..((((((((.(((((.....((((.(((........(((((..((((........)))).....))))).((((......)))).)))))))))))).))))))))..)..... ( -38.90) >consensus GCAGUUAUGGCAGCUAAAGAUCCUUUGGCGUGGACAAAU______GUAUGCCAUA_AAUGGGCGUAGGAUGCUCC____UUCGAUGGAAACUGGCCAUCUUUUGCUGCCAUCUCAAUUU ...(..((((((((.(((((.....((((((((.((.((......)).)))))).....(((((.....)))))...........(....)..))))))))).))))))))..)..... (-31.40 = -31.36 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:35 2006