
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
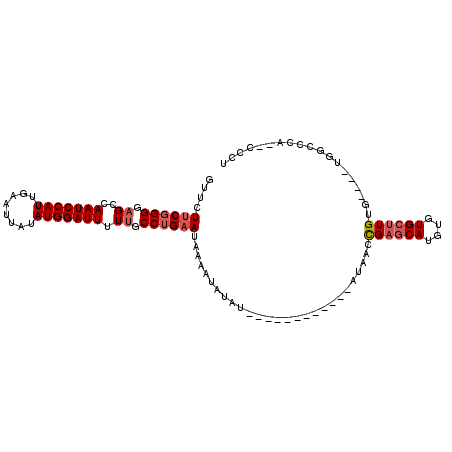
| Location | 13,647,352 – 13,647,443 |
| Length | 91 |
| Max. P | 0.589075 |

| Location | 13,647,352 – 13,647,443 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -16.57 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13647352 91 + 23771897 GUUCUUCGGGGAGCCAAUCCAUUGAAUUAUAUGGAUUUUUGCCUGAAUAAAAUAUAU------------AUAACCGAGCAUGUGUGCUUGUG----UGGCCCA--CCCU .......((((.((((((((((........)))))))....................------------...((((((((....)))))).)----)))).).--))). ( -24.60) >DroSec_CAF1 54754 95 + 1 GUUCUUCGGGGAGCCAAUCCAUUGAAUUAUAUGGAUUUUUGCCUGAAUAAAAUAUAU------------AUAACCGAGCAUGUGUGCUUGUGAGGGUGGUUCA--CCCU .......(((((((((((((((........)))))))...((((..((((..(((((------------((........))))))).))))..))))))))).--))). ( -28.20) >DroSim_CAF1 64898 95 + 1 GUUCUUCGGGGAGCCAAUCCAUUGAAUUAUAUGGAUUUUUGCCUGAAUAAAAUAUAU------------AUAACCGAGCAUGUGUGCUUGUGUGGGUGGUUCA--CCCU .......(((((((((.(((((....(((((((.(((((.........))))).)))------------)))).((((((....)))))).))))))))))).--))). ( -29.30) >DroEre_CAF1 57463 96 + 1 GUUCUUCGGGG-GCCAAUCCAUUGAAUUAUAUGGAUUUUUGCCUGAAUAAAAUAUAUACG------AGUAUAGCCGAGCAUGUGUGCUUGUG----UGGGCCA--CCCU .......((((-((((((((((........))))))).................((((((------((((((((...))...))))))))))----)))))).--))). ( -31.60) >DroYak_CAF1 62535 97 + 1 GUUCUUCGGGGAGCCAAUCCAUUGAAUUAUAUGGAUUUUUGCCUGAAUAAAAUAUAUACAUUCACUCGUAUAACCGAGCAUGUGUGCUUG----------CCA--CCCU ....((((((.((..(((((((........))))))).)).)))))).......(((((........)))))..((((((....))))))----------...--.... ( -22.70) >DroAna_CAF1 92256 93 + 1 GUGCUCC-GGGAGCCAAUCCAUUGAAUUAUAUGGAUUUUUGCCCGAAUAAAGUAUAC------------A--UCUGGGCAUGUGUGGGUGCCAC-CCGGCCCAAGCCCA (((((.(-(((....(((((((........)))))))....)))).....)))))..------------.--...((((.((.(((((.....)-)).)).)).)))). ( -30.90) >consensus GUUCUUCGGGGAGCCAAUCCAUUGAAUUAUAUGGAUUUUUGCCUGAAUAAAAUAUAU____________AUAACCGAGCAUGUGUGCUUGUG____UGGCCCA__CCCU ....((((((.((..(((((((........))))))).)).))))))...........................((((((....))))))................... (-16.57 = -17.27 + 0.69)


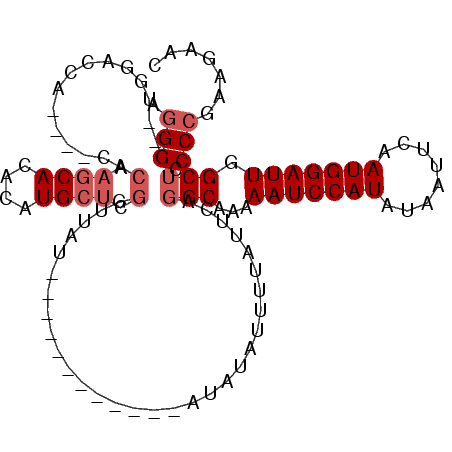
| Location | 13,647,352 – 13,647,443 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -13.40 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13647352 91 - 23771897 AGGG--UGGGCCA----CACAAGCACACAUGCUCGGUUAU------------AUAUAUUUUAUUCAGGCAAAAAUCCAUAUAAUUCAAUGGAUUGGCUCCCCGAAGAAC .(((--.(((((.----............(((..(..((.------------........))..)..)))..(((((((........)))))))))))))))....... ( -24.80) >DroSec_CAF1 54754 95 - 1 AGGG--UGAACCACCCUCACAAGCACACAUGCUCGGUUAU------------AUAUAUUUUAUUCAGGCAAAAAUCCAUAUAAUUCAAUGGAUUGGCUCCCCGAAGAAC ((((--((...)))))).....(((....)))((((....------------(((.....)))...(((...(((((((........))))))).)))..))))..... ( -20.80) >DroSim_CAF1 64898 95 - 1 AGGG--UGAACCACCCACACAAGCACACAUGCUCGGUUAU------------AUAUAUUUUAUUCAGGCAAAAAUCCAUAUAAUUCAAUGGAUUGGCUCCCCGAAGAAC .(((--((...)))))......(((....)))((((....------------(((.....)))...(((...(((((((........))))))).)))..))))..... ( -21.00) >DroEre_CAF1 57463 96 - 1 AGGG--UGGCCCA----CACAAGCACACAUGCUCGGCUAUACU------CGUAUAUAUUUUAUUCAGGCAAAAAUCCAUAUAAUUCAAUGGAUUGGC-CCCCGAAGAAC .(((--(((((..----....((((....)))).)))))....------.................(((...(((((((........))))))).))-))))....... ( -23.90) >DroYak_CAF1 62535 97 - 1 AGGG--UGG----------CAAGCACACAUGCUCGGUUAUACGAGUGAAUGUAUAUAUUUUAUUCAGGCAAAAAUCCAUAUAAUUCAAUGGAUUGGCUCCCCGAAGAAC .(((--..(----------(..((......(((((......)))))(((((.........)))))..))...(((((((........))))))).))..)))....... ( -27.90) >DroAna_CAF1 92256 93 - 1 UGGGCUUGGGCCGG-GUGGCACCCACACAUGCCCAGA--U------------GUAUACUUUAUUCGGGCAAAAAUCCAUAUAAUUCAAUGGAUUGGCUCCC-GGAGCAC ...(((((((((((-(.....))).....(((((..(--(------------(.......)))..)))))..(((((((........))))))))))))..-.)))).. ( -33.40) >consensus AGGG__UGGACCA____CACAAGCACACAUGCUCGGUUAU____________AUAUAUUUUAUUCAGGCAAAAAUCCAUAUAAUUCAAUGGAUUGGCUCCCCGAAGAAC .(((...............(.((((....)))).)...............................(((...(((((((........))))))).))).)))....... (-13.40 = -14.23 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:31 2006