
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,639,812 – 13,639,965 |
| Length | 153 |
| Max. P | 0.978917 |

| Location | 13,639,812 – 13,639,926 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -30.08 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
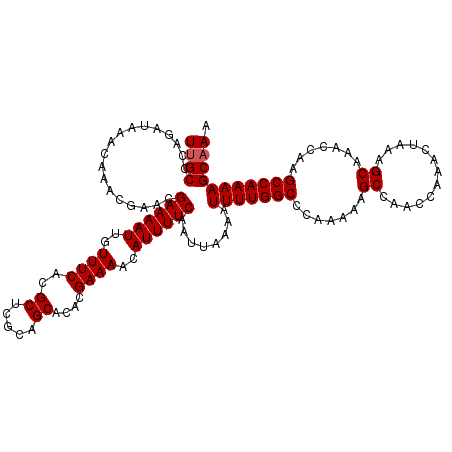
>3L_DroMel_CAF1 13639812 114 + 23771897 CAAUGUGAGAAACGUUUUCGGUGCUCCGCACUGCUUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCG ..(..((((((.((((((((((((...))))).......((((((((..(...(((...........)))...)..))))))))........)))))))))))))..)...... ( -32.30) >DroSec_CAF1 47328 114 + 1 CAAUGUGAGAAACGUUUUCGAUGCUCCGCACUGCUUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCG ..(..((((((.((((((((((.....(((......)))((((((((..(...(((...........)))...)..)))))))).....))))))))))))))))..)...... ( -32.10) >DroSim_CAF1 57299 114 + 1 CAAUGUGAGAAACGUUUUCGGUGCUCCGCACUGCUUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCG ..(..((((((.((((((((((((...))))).......((((((((..(...(((...........)))...)..))))))))........)))))))))))))..)...... ( -32.30) >DroEre_CAF1 49719 114 + 1 CAAUGUGAGAAACGCUUUCGGUGCUACGCGCUGCUUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCG ..(..((((((..((...((((((...))))))....))((((((((..(...(((...........)))...)..))))))))...............))))))..)...... ( -28.20) >DroYak_CAF1 54460 114 + 1 CAAUGUGAGAAACGCUUUUGGUGCUCCGCAGUGCUUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCG ......(((...(((.....))))))(((((..(.....((((((((..(...(((...........)))...)..))))))))........((((....))))..)..))))) ( -29.70) >consensus CAAUGUGAGAAACGUUUUCGGUGCUCCGCACUGCUUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCG ..(..((((((.((((((((((.....(((......)))((((((((..(...(((...........)))...)..)))))))).....))))))))))))))))..)...... (-30.08 = -30.00 + -0.08)



| Location | 13,639,846 – 13,639,965 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 99.16 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -29.18 |
| Energy contribution | -29.02 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13639846 119 + 23771897 UUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCGAGCGUGAAACAAUUUUCGUUCGUUUGUUUAUCUGGGCAA .....((((((((..(...(((...........)))...)..)))))))).............(((((..(((.((.(((((((.(((....))).)))))))..)).)))..))))). ( -29.20) >DroSec_CAF1 47362 119 + 1 UUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCGAGCGUGAAACAAUUUUCGUUCGUUUGUUUAUCUGGGCAA .....((((((((..(...(((...........)))...)..)))))))).............(((((..(((.((.(((((((.(((....))).)))))))..)).)))..))))). ( -29.20) >DroSim_CAF1 57333 119 + 1 UUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCGAGCGUGAAACAAUUUUCGUUCGUUUGUUUAUCUGGGCAA .....((((((((..(...(((...........)))...)..)))))))).............(((((..(((.((.(((((((.(((....))).)))))))..)).)))..))))). ( -29.20) >DroEre_CAF1 49753 119 + 1 UUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCGAGCGUGAAACAAUUUUCGUUCGUCUGUUUAUCUGGGCAA .....((((((((..(...(((...........)))...)..)))))))).............(((((..(((.((.(((((((.(((....))).)))))))..)).)))..))))). ( -29.10) >DroYak_CAF1 54494 119 + 1 UUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCGAGCGUGAAACAAUUUUCGUUCGUCUGUUUAUCUGGGCGA .....((((((((..(...(((...........)))...)..))))))))........((((((..((((((((.......))))))))..))))))..((((((........)))))) ( -29.40) >consensus UUUGCUUUUGGCUUGGUUUGCUUUAGUUUGGUUGGCUUUUUGGGCCAAAAUUUUAAUUGAAAAUGUUUUCGUGUGCUGCGAGCGUGAAACAAUUUUCGUUCGUUUGUUUAUCUGGGCAA .....((((((((..(...(((...........)))...)..)))))))).............(((((..(((.((.(((((((.(((....))).)))))))..)).)))..))))). (-29.18 = -29.02 + -0.16)



| Location | 13,639,846 – 13,639,965 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 99.16 |
| Mean single sequence MFE | -16.10 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13639846 119 - 23771897 UUGCCCAGAUAAACAAACGAACGAAAAUUGUUUCACGCUCGCAGCACACGAAAACAUUUUCAAUUAAAAUUUUGGCCCAAAAAGCCAACCAAACUAAAGCAAACCAAGCCAAAAGCAAA ((((..................((((((..((((..((.....))....))))..))))))........(((((((.......((.............)).......))))))))))). ( -16.36) >DroSec_CAF1 47362 119 - 1 UUGCCCAGAUAAACAAACGAACGAAAAUUGUUUCACGCUCGCAGCACACGAAAACAUUUUCAAUUAAAAUUUUGGCCCAAAAAGCCAACCAAACUAAAGCAAACCAAGCCAAAAGCAAA ((((..................((((((..((((..((.....))....))))..))))))........(((((((.......((.............)).......))))))))))). ( -16.36) >DroSim_CAF1 57333 119 - 1 UUGCCCAGAUAAACAAACGAACGAAAAUUGUUUCACGCUCGCAGCACACGAAAACAUUUUCAAUUAAAAUUUUGGCCCAAAAAGCCAACCAAACUAAAGCAAACCAAGCCAAAAGCAAA ((((..................((((((..((((..((.....))....))))..))))))........(((((((.......((.............)).......))))))))))). ( -16.36) >DroEre_CAF1 49753 119 - 1 UUGCCCAGAUAAACAGACGAACGAAAAUUGUUUCACGCUCGCAGCACACGAAAACAUUUUCAAUUAAAAUUUUGGCCCAAAAAGCCAACCAAACUAAAGCAAACCAAGCCAAAAGCAAA ((((..................((((((..((((..((.....))....))))..))))))........(((((((.......((.............)).......))))))))))). ( -16.36) >DroYak_CAF1 54494 119 - 1 UCGCCCAGAUAAACAGACGAACGAAAAUUGUUUCACGCUCGCAGCACACGAAAACAUUUUCAAUUAAAAUUUUGGCCCAAAAAGCCAACCAAACUAAAGCAAACCAAGCCAAAAGCAAA ..((..................((((((..((((..((.....))....))))..))))))........(((((((.......((.............)).......)))))))))... ( -15.06) >consensus UUGCCCAGAUAAACAAACGAACGAAAAUUGUUUCACGCUCGCAGCACACGAAAACAUUUUCAAUUAAAAUUUUGGCCCAAAAAGCCAACCAAACUAAAGCAAACCAAGCCAAAAGCAAA ((((..................((((((..((((..((.....))....))))..))))))........(((((((.......((.............)).......))))))))))). (-15.56 = -15.76 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:28 2006