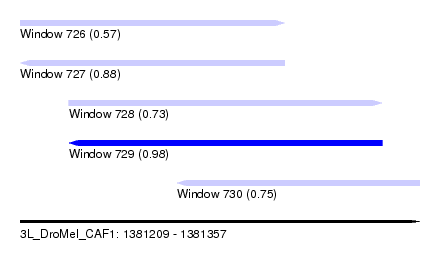
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,381,209 – 1,381,357 |
| Length | 148 |
| Max. P | 0.982950 |

| Location | 1,381,209 – 1,381,307 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -28.13 |
| Energy contribution | -27.68 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381209 98 + 23771897 ---G--UCGAU--CUGCUAUCACAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGGUGAA ---.--.....--......((((.(((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).....)))). ( -30.70) >DroPse_CAF1 60582 98 + 1 UUUAACGUGAG--CACUUUCCAUAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUG----- .....(....)--...((((((((((((((........))))...(((.(((((((((((....)))))..)))))).....))).))))))))))....----- ( -32.80) >DroEre_CAF1 37949 98 + 1 ---G--CCGGA--CCCCUAUCACAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAAUGGUGGA ---(--(((((--..((((((((...............)))).))))...))))))(((((.((....)).(((((.......)))))..))))).......... ( -34.26) >DroYak_CAF1 39547 98 + 1 ---G--GCAAU--CCCCUAUCACAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAGUAGUGAA ---.--.....--......((((.(((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).....)))). ( -29.60) >DroAna_CAF1 38388 94 + 1 ---G--GCAGACGCCAUUUUCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACU------ ---(--((....))).........(((((((.(((((........))))).((((.((..((((((.....))))))..))...)))))))))))....------ ( -33.10) >DroPer_CAF1 61615 98 + 1 UUUGACGUGAG--CACUUUCCAUAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUG----- ..((.(....)--)).((((((((((((((........))))...(((.(((((((((((....)))))..)))))).....))).))))))))))....----- ( -33.80) >consensus ___G__GCGAA__CCCCUAUCACAUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAACUG_____ ........................(((((((.(((((........)))))(((((.......)))))....(((((.......)))))))))))).......... (-28.13 = -27.68 + -0.44)

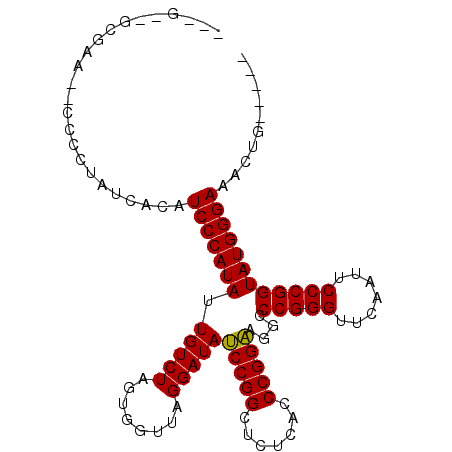
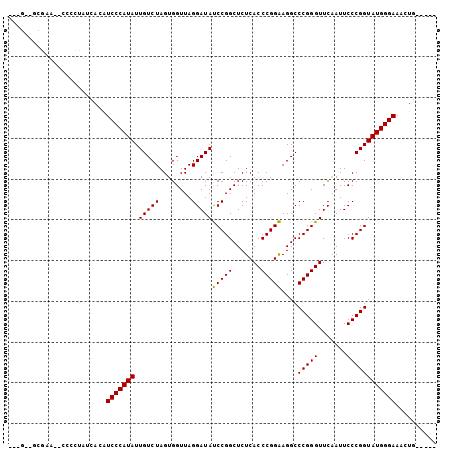
| Location | 1,381,209 – 1,381,307 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.02 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381209 98 - 23771897 UUCACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUGUGAUAGCAG--AUCGA--C--- .....(((((.((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))))))).......--.....--.--- ( -31.10) >DroPse_CAF1 60582 98 - 1 -----CAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUAUGGAAAGUG--CUCACGUUAAA -----((.(((((....((((...((..(((((((...)))))))..)).))))((((((((..............))))))))))))).))--........... ( -34.44) >DroEre_CAF1 37949 98 - 1 UCCACCAUUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUGUGAUAGGGG--UCCGG--C--- .........(((((.....)))))((..((((((.....))))))..))(((((((.(((((..(((....(.....)....))).))))))--)))))--)--- ( -35.70) >DroYak_CAF1 39547 98 - 1 UUCACUACUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUGUGAUAGGGG--AUUGC--C--- .........(((((...((((...((..((((((.....))))))..)).))))((((((((..............))))))))....))))--)....--.--- ( -32.34) >DroAna_CAF1 38388 94 - 1 ------AGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGAAAAUGGCGUCUGC--C--- ------....(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....))))))).........(((....))--)--- ( -32.10) >DroPer_CAF1 61615 98 - 1 -----CAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUAUGGAAAGUG--CUCACGUCAAA -----((.(((((....((((...((..(((((((...)))))))..)).))))((((((((..............))))))))))))).))--........... ( -34.44) >consensus _____CAGUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAUGUGAAAAGGG__AUCGC__C___ ..........(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....)))))))........................ (-29.38 = -29.02 + -0.36)

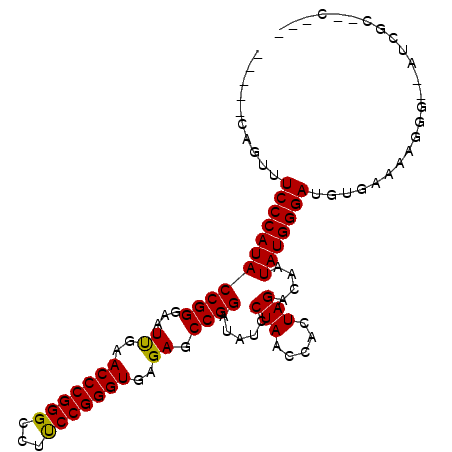
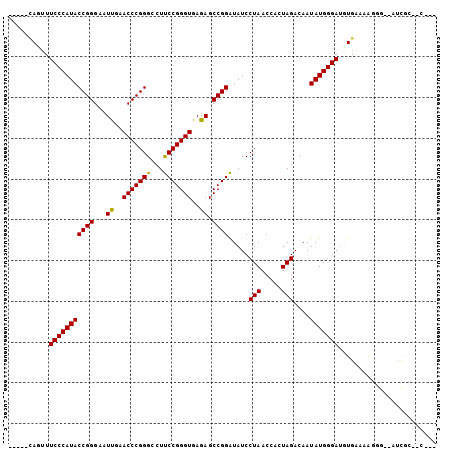
| Location | 1,381,227 – 1,381,343 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -25.70 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381227 116 + 23771897 CCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGGUGAAGAAAU----AUUCUUUUUAGAAAUUUUCACUUUAAUUAUA ((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))..((.((((((((...----.((((....)))).)))))))).))...... ( -33.40) >DroSec_CAF1 36794 116 + 1 CCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAUAUGGUGAAGAAAU----AUUCUUUUUAGAAACUUUUACUUUAAUUUAA .(((((((.((((.(((..(....(((((((..((.....))..).))))))....)..))).)).)).)))))))((((((((.----.((((....))))...))).)))))...... ( -29.70) >DroEre_CAF1 37967 119 + 1 CCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAAUGGUGGAAAUGUUUAUUUUCUUUUGAGAAACUUAAAAUAAAA-UUUA ((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))..................(((((.(((((((...))))))).))))-)... ( -29.00) >DroWil_CAF1 15473 102 + 1 CCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACUU------------------UUUUUCCCUCGUAUUAUUAUAGUAUUA ...(((((((..((((((.(.((...((((.((..((((((.....))))))..))...))))...((((((...------------------..)))))))).)))))))))))))).. ( -27.90) >DroYak_CAF1 39565 107 + 1 CCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAGUAGUGAAAAUGU----UUUCUUUUAAGAAA--------ACAA-GUUA ((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))).............(((----(((((....)))))--------))).-.... ( -32.50) >consensus CCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAAUGGUGAAAAAAU____AUUCUUUUUAGAAACUUUUACUAUAAUUUUA ((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))................................................... (-25.70 = -25.70 + -0.00)

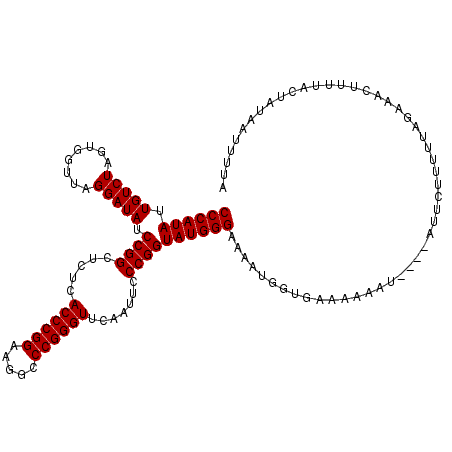
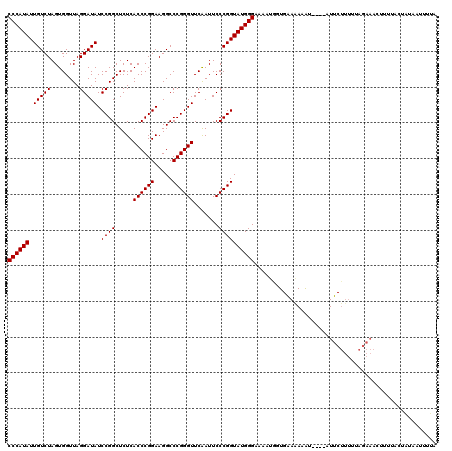
| Location | 1,381,227 – 1,381,343 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -26.12 |
| Energy contribution | -25.96 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381227 116 - 23771897 UAUAAUUAAAGUGAAAAUUUCUAAAAAGAAU----AUUUCUUCACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGG ..........(((((...((((....)))).----.....)))))......((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))) ( -29.90) >DroSec_CAF1 36794 116 - 1 UUAAAUUAAAGUAAAAGUUUCUAAAAAGAAU----AUUUCUUCACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGG ..................((((....)))).----.........(((((((......((((...((..((((((.....))))))..)).)))).....(((.....)))..))))))). ( -26.20) >DroEre_CAF1 37967 119 - 1 UAAA-UUUUAUUUUAAGUUUCUCAAAAGAAAAUAAACAUUUCCACCAUUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGG ....-............(((((....)))))....................((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))) ( -27.00) >DroWil_CAF1 15473 102 - 1 UAAUACUAUAAUAAUACGAGGGAAAAA------------------AAGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGG .....(((((.........((((((..------------------...))))))...((((...((..((((((.....))))))..)).))))....................))))). ( -31.50) >DroYak_CAF1 39565 107 - 1 UAAC-UUGU--------UUUCUUAAAAGAAA----ACAUUUUCACUACUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGG ....-.(((--------(((((....)))))----))).............((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))) ( -32.10) >consensus UAAAAUUAUAGUAAAAGUUUCUAAAAAGAAA____ACAUCUUCACCAUUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGG ...................................................((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))) (-26.12 = -25.96 + -0.16)

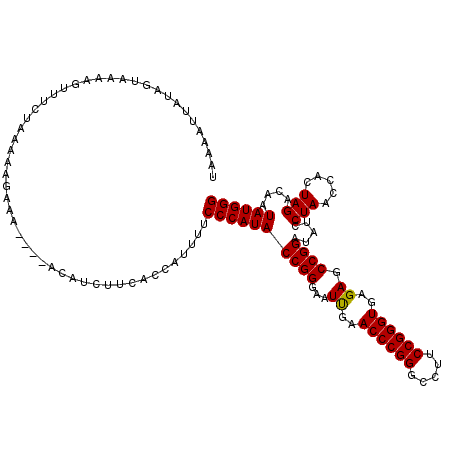
| Location | 1,381,267 – 1,381,357 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 78.58 |
| Mean single sequence MFE | -12.93 |
| Consensus MFE | -9.88 |
| Energy contribution | -9.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1381267 90 - 23771897 GACCAAUAAAUAAAUAUAAUUAAAGUGAAAAUUUCUAAAAAGAAU----AUUUCUUCACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUC ..((....................(((((...((((....)))).----.....)))))((.((((((.....)))))))).....))...... ( -14.30) >DroSec_CAF1 36834 90 - 1 GACCAAUAAAUAAAUUAAAUUAAAGUAAAAGUUUCUAAAAAGAAU----AUUUCUUCACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUC ..((............................((((....)))).----.....((((....((((((.....))))))))))...))...... ( -10.90) >DroEre_CAF1 38007 93 - 1 GAUCAUUAAUUACAUAAA-UUUUAUUUUAAGUUUCUCAAAAGAAAAUAAACAUUUCCACCAUUUUCCCAUACCGGGAAUUGAACCCGGGCCUUC ..................-............(((((....)))))..........................(((((.......)))))...... ( -10.70) >DroYak_CAF1 39605 80 - 1 GAUCAAUAAU-AAAUAAC-UUGU--------UUUCUUAAAAGAAA----ACAUUUUCACUACUUUCCCAUACCGGGAAUUGAACCCGGGCCUUC ..........-.......-.(((--------(((((....)))))----)))...................(((((.......)))))...... ( -15.80) >consensus GACCAAUAAAUAAAUAAA_UUAAAGU_AAAGUUUCUAAAAAGAAA____ACAUCUUCACCAUAUUCCCAUACCGGGAAUUGAACCCGGGCCUUC ................................((((....))))...........................(((((.......)))))...... ( -9.88 = -9.88 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:19 2006