
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,638,824 – 13,638,930 |
| Length | 106 |
| Max. P | 0.783352 |

| Location | 13,638,824 – 13,638,930 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
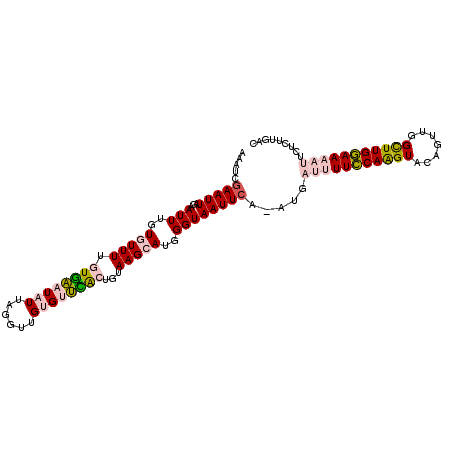
>3L_DroMel_CAF1 13638824 106 + 23771897 AAAUCGAAUUAGAAUUUGUGUUUUGUGAAUAUUAGGUUGUGUUCACUGUAAGCAUGGGUAAUUCA-AUGAUUUUCCAAGUACAGUUAGCUUGGAAAAUUCUCUUGAC .....(((((...(((..(((((.((((((((......))))))))...)))))..)))))))).-..((((((((((((.......))))))))))))........ ( -31.20) >DroPse_CAF1 30666 105 + 1 AAAUCGAAUUAAAAUUUGUAUUUCAUUGUUUUU--UUCGUGUCGGGUGUAAGCAUGGGUAAUUCGGAGCGCUUUUCAGGUACAGUCGGUUUGAAAUAAUUUCUUUUC (((((((((....))))).)))).........(--((((..((((.(((..((.(.((....)).).))(((.....)))))).))))..)))))............ ( -13.50) >DroSim_CAF1 56302 106 + 1 AAAUCGAAUUAGAAUUUGUGUUUUGUGAAUAUUAGGUUGUGUUCACUGUAAGCAUGGGUAAUUGA-AUGAUUUUCCAAGUACAGUUAGCUUGGAAAAUUCUCUUGAC ...((((......(((..(((((.((((((((......))))))))...)))))..)))....((-..((((((((((((.......)))))))))))).)))))). ( -29.90) >DroEre_CAF1 48725 106 + 1 AAAUCGAAUUAGAAUUUGUGUUUUGUGAAUAUUAGAUUGUGUUCACUGUAAGCAUGGGUAAUUCA-AUUAUUUUCCAAGUACGUUUGGCUUGGAAAAUUCUCUUGAC .....(((((...(((..(((((.((((((((......))))))))...)))))..)))))))).-...(((((((((((.......)))))))))))......... ( -30.00) >DroYak_CAF1 53475 106 + 1 AAAUCGAAUUAGAAUUUGUGUUUUGUGAAUAUUAGGUUGUGUUUACUGUAAGCAUGGGUAAUUCA-AUGAUUUUCCAAGUACAGUUGGCUUGGAAAAUUCUCUUGAC .....(((((...(((..(((((.((((((((......))))))))...)))))..)))))))).-..((((((((((((.......))))))))))))........ ( -29.10) >DroPer_CAF1 30634 105 + 1 AAAUCGAAUUAAAAUUUGUAUUUCAUUGUUUUU--UUCGUGUCGGGUGUAAGCAUGGGUAAUUCGGAGCGCUUUUCAGGUACAGUCGGUUUGAAAUAAUUUUUUUUC .....(((..((((((................(--((((..((((.(((..((.(.((....)).).))(((.....)))))).))))..))))).))))))..))) ( -14.13) >consensus AAAUCGAAUUAGAAUUUGUGUUUUGUGAAUAUUAGGUUGUGUUCACUGUAAGCAUGGGUAAUUCA_AUGAUUUUCCAAGUACAGUUGGCUUGGAAAAUUCUCUUGAC .....(((((...(((..(((((.((((((((......))))))))...)))))..)))))))).....(((((((((((.......)))))))))))......... (-19.00 = -19.42 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:25 2006