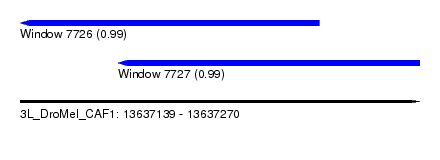
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,637,139 – 13,637,270 |
| Length | 131 |
| Max. P | 0.990626 |

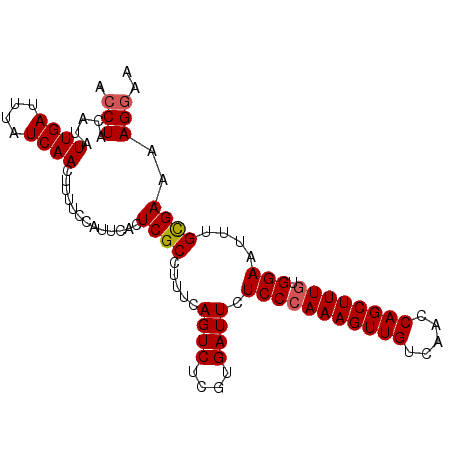
| Location | 13,637,139 – 13,637,237 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.53 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13637139 98 - 23771897 CUCGCCUUUCAGUCUCGUGAUUCUCCAAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAUAGGAAAAA--AACCUUAAAUUCUAAUCGGAUUUACAUUU ......((((....(((..(...(((((((((((.....))))))).))))..)..)))....))))..--.....(((((((.....)))))))..... ( -20.80) >DroSec_CAF1 44595 98 - 1 CUCGCCUUUCAGUCUCGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAAAAA--AACCUUAAAUUCUAAUCGGAUUUACAUUU ....(((((.....(((..(...(((((((((((.....)))))))).)))..)..)))))))).....--.....(((((((.....)))))))..... ( -24.00) >DroSim_CAF1 54609 98 - 1 CUCGCCUUUCAGUCUCGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAAAAA--AACCUUAAAUUCUAAUCGGAUUUACAUUU ....(((((.....(((..(...(((((((((((.....)))))))).)))..)..)))))))).....--.....(((((((.....)))))))..... ( -24.00) >DroEre_CAF1 47007 98 - 1 CUCGCUCCUCAGUCUGGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAACAA--AACCUUAAAUUCUAAUCGGAUUUACAUUU .((((.....((((....)))).(((((((((((.....)))))))).)))....)))).((((.....--..))))((((((.....))))))...... ( -22.00) >DroYak_CAF1 51749 100 - 1 CUCGCCUUUCAGUCUGAUGAUUCUCCCAAAGUUGUCAGCCAGCUUUGUGGAAUUUGUGAAAAGCAAAAAAGAACCUUAAAUUCUAAUCGGAUUUACAUUU ...((.((((((((....)))..(((((((((((.....)))))))).))).....))))).))............(((((((.....)))))))..... ( -21.80) >consensus CUCGCCUUUCAGUCUCGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAAAAA__AACCUUAAAUUCUAAUCGGAUUUACAUUU .((((.....((((....)))).(((((((((((.....)))))))).)))....)))).................(((((((.....)))))))..... (-19.28 = -19.32 + 0.04)

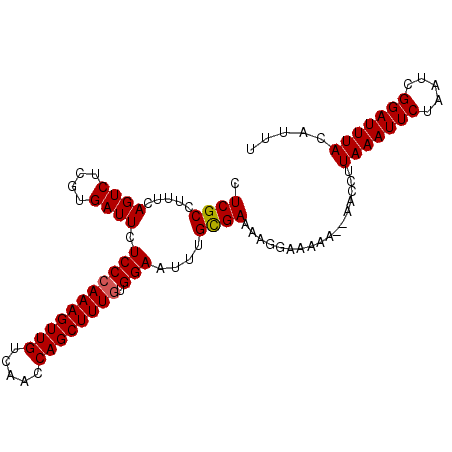
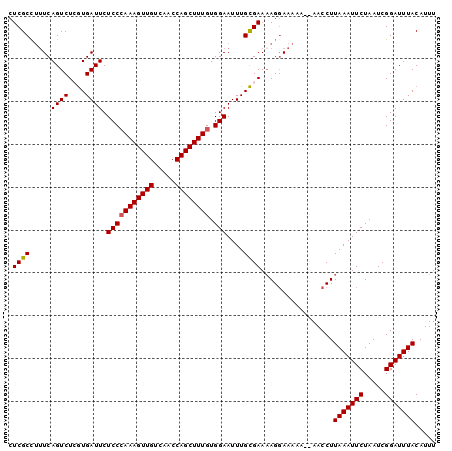
| Location | 13,637,171 – 13,637,270 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 94.95 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13637171 99 - 23771897 ACCUACAUAUUGAUUUAUCAACUUUUCCAUUCACUCGCCUUUCAGUCUCGUGAUUCUCCAAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAUAGGAA .((((....((((....))))..........................(((..(...(((((((((((.....))))))).))))..)..))).)))).. ( -19.90) >DroSec_CAF1 44627 99 - 1 ACCUAUAUAUUGAUUUAUCAACUUUUCCAUUCACUCGCCUUUCAGUCUCGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAA .........((((....))))................(((((.....(((..(...(((((((((((.....)))))))).)))..)..)))))))).. ( -21.40) >DroSim_CAF1 54641 99 - 1 ACCUACAUAUUGAUUUAUCAACUUUUCCAUUCACUCGCCUUUCAGUCUCGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAA .........((((....))))................(((((.....(((..(...(((((((((((.....)))))))).)))..)..)))))))).. ( -21.40) >DroEre_CAF1 47039 99 - 1 ACCUACAUAUUGAUUUAUCAACUUUUACAUUCACUCGCUCCUCAGUCUGGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAA .(((.....((((....)))).............((((.....((((....)))).(((((((((((.....)))))))).)))....))))..))).. ( -21.40) >DroYak_CAF1 51783 99 - 1 ACCUACAUAUUGAUUUAUCAACUUUUCCAUUCACUCGCCUUUCAGUCUGAUGAUUCUCCCAAAGUUGUCAGCCAGCUUUGUGGAAUUUGUGAAAAGCAA .........((((....))))...............((.((((((((....)))..(((((((((((.....)))))))).))).....))))).)).. ( -19.20) >consensus ACCUACAUAUUGAUUUAUCAACUUUUCCAUUCACUCGCCUUUCAGUCUCGUGAUUCUCCCAAAGUUGUCAACCAGCUUUGUGGAAUUUGCGAAAAGGAA .(((.....((((....)))).............((((.....((((....)))).(((((((((((.....)))))))).)))....))))..))).. (-18.28 = -18.52 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:21 2006